Molecular Biology: From DNA to RNA to Protein
4 DNA Packaging in Eukaryotes
4.1 Introduction: “Inner Life of the Genome”
The size of the genome in one of the most well-studied prokaryotes, E.coli, is 4.6 million base pairs (approximately 1.1 mm if cut and stretched out).
Let’s do a rough calculation about how much DNA a eukaryotic cell has.
Assume
- A somatic cell has 2 full copies of every chromosome. The total number of base pairs for ALL chromosomes combined is 6 X 109 bp.
- We know the length of each base pair is 0.34nm (see DNA structure chapter!)
If you were to take the DNA of all the chromosomes, stretch it out, and lay them end to end then what will the total length of ALL the DNA in the cell be?
Hopefully, you got to a number of 2.0 meters! Now consider the size of the eukaryotic nucleus which ranges from 2- 10 microns! For a good frame of reference, a single grain of salt is 100 microns which is still 10 times larger than the upper range for nucleus size!
This results in an engineering problem that cells need to solve.
Problem 1: How does all of the DNA fit inside a bacterial cell and even more amazingly inside the nucleus of a eukaryotic cell?
At the same time consider all the different cells in the body. The human body contains approximately many different cell types, but each cell type shares the same genomic sequence. In spite of having the same genetic code, cells not only develop into distinct types from this same sequence but also maintain the same cell type over time and across divisions.
They are also specialized in their function- liver cells for example produce enzymes that help with detoxification but they do not produce antibodies, that is the job of a different cell type.
These cells have different expression patterns due to the temporal and spatial regulation of genes. (See video below for a good analogy).
This leads to the second problem to contend with- once the genome has been folded and all packaged up how can we open small sections of it so the machinery to read and express the information coded within the genes can gain access?
The epigenome (“epi” means above in Greek, so epigenome means above genome) is the set of chemical modifications or marks that influence gene expression and are transferred across cell divisions and, in some limited cases, across generations of organisms.
In this chapter, you will learn about the solution to the ‘Packaging Problem’ and how cells regulate access to genes.
The solution involves interactions of DNA with specific proteins, leading to the formation of a nucleoprotein complex called CHROMATIN. We will focus exclusively on Eukaryotic chromatin, although bacterial chromosomes (which are circular) also coil and supercoil with the help of proteins.
Learning Objectives
Levels 1 and 2 are factual information, and knowledge-based. Level up indicated by the “Target” symbol is the goal.
When you have mastered the information in this chapter, you should be able to:
Level 1 and 2 (Knowledge and Comprehension)
- Draw/Label nucleosomes in a 10nm fiber to identify core histones, linker DNA, and core DNA.
- Outline the steps by which a nucleosome is assembled.
- List the five major types of histone proteins, and describe what role each of them plays in the nucleosome
- Explain what form of chromatin is present during interphase.
- Distinguish among the levels of chromatin packing in a eukaryotic chromosome
- Define the roles of modifying amino acid side chains in altering nucleosome structure and how the structure of chromatin can be used for regulating gene expression.
- o What modifications would lead to activation of gene expression?
⊕ Level Up (Application, Analysis, Synthesis)
- Analyze/Predict/Interpret experimental data connected with DNA organization into chromatin.
- Analyze/Predict/Interpret experimental data connecting chromatin structure with gene expression.
- Predict how mutations in key histones can alter chromatin structure /gene expression.
- Outline an experiment to purify histones from chromatin.
4.2 Types of Chromatin in Eukaryotic Cells
When we visualize chromosomes we usually think of the characteristic highly condensed structures (X-shaped). DNA in this form exists only transiently for a brief phase in Mitosis, where it is maximally condensed.
These discrete packets are ideal for the job at hand which is to accurately separate and segregate to opposite poles of the cell. The majority of the time a cell is in fact in interphase and not dividing.
During Interphase which can be further subdivided into Gap 1-G1, S-phase, and G2] cells are transcribing (RNA) and translating (making proteins) sometimes on demand and depending on the needs of the cell.
Not all parts of the genome however are active. During interphase (Figure 4.1 below) chromatin exists in various states of condensation called heterochromatin and euchromatin respectively.
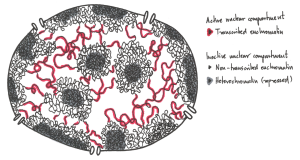
The transition between these chromatin forms involves changes in the amounts and types of proteins bound to the chromatin that can occur during gene regulation, i.e. when genes are turned on or off.
Experiments to be described later showed that active genes tend to be in the more dispersed euchromatin where enzymes of replication and transcription have easier access to the DNA. Transcriptionally inactive genes are heterochromatic, obscured by additional chromatin proteins present in heterochromatin.
TERMINOLOGY CHECK!
“Before we get started, it is absolutely vital that we briefly review what you already know about the structure of the eukaryotic genome from previous courses. The terminology is very confusing, as many of the terms are similar, so it’s worth taking a moment to go over it. An overview that helps you connect the terms together will be beneficial for you to refer to later. The eukaryotic genome is made up of a number of chromosomes. In diploid organisms such as ourselves, there are two copies of each chromosome (for comparison, haploid organisms only have one copy of each chromosome). As a reminder, humans have 23 pairs of chromosomes. These “pairs” of chromosomes are similar in that they have all of the same genes in the same order, but they often carry different versions of the genes, which are known as alleles. One set of 23 chromosomes comes from our egg-bearing biological parent, and the other set we acquire from our sperm-bearing biological parent.
During most of the cell’s life, each of these chromosomes will be made of a single chromatid, and that chromatid will exist as chromatin. Chromatin is a complex of DNA and proteins that helps keep the DNA organized inside the nucleus”. (4)
Exercises
4.3 Chromatin Organization
Chromatin consists of DNA combined with two classes of proteins, which are known as histones and nonhistone chromatin-associated proteins. We will devote much of our time to Histones as they are responsible for the first and fundamental unit of packaging.
4.3.1 Histones
Histones are a set of proteins that interact strongly, but reversibly, with DNA. They are found in all eukaryotes (i.e., organisms with a nucleus and nuclear envelope), and even Archaea, but not bacteria.
There are 5 major classes of histones: H1, H2A, H2B, H3, and H4. In keeping with their role in associating with DNA, Histones are considered to be “basic” proteins due to the overall positive charge. This charge comes from the protein containing many lysine and arginine amino acids. Their positively charged side chains enable these amino acids to bind to the acidic, negatively charged phosphodiester backbone of double-helical DNA.
The functional importance of histones is reflected in how well conserved they are in different species. Histone H4 has an extremely well conserved amino acid sequence among eukaryotes, as does H3. For instance, the H4 in cows differs from H4 in peas by only 2 amino acids! H2A and H2B have moderately conserved amino acid sequences while H1 has the most sequence variation. The amino acid sequence of H1 has the most variation varies between and even within species.
4.3.2 First level of packing- “Beads on a String” structure
How do the histones help organize DNA? Here looking at the experiments to identify and isolate these proteins becomes helpful. Investigators started by gently disrupting cell nuclei to extract the chromatin using salts. The salts helped “unfold” the chromatin and they were able to visualize what it looked like using an electron microscope. The resulting image looked like “ beads on a string ”
Here the string is DNA, which is wrapped around nucleosome core particles, the beads. To determine what makes up this nucleosome (“bead”) scientists used enzymes called nucleases.
Nucleases cut DNA by breaking phosphodiester bonds between nucleotides. The particular enzyme used is only able to cut exposed DNA (“the string” between the “beads” )
When this was carried out for a SHORT time, the nucleosome particles were released. Electrophoresis of DNA extracted from digests of nucleosome beads-on-a-string preparations generated DNA fragments about 200 base pairs in length.
In contrast, the digestion of naked DNA (not associated with proteins) yielded a continuous smear of randomly sized fragments.
These results suggested that the binding of proteins to DNA in chromatin protects regions of the DNA from nuclease digestion so that the enzyme can attack DNA only at sites separated by approximately 200 base pairs.
This revealed the basic unit of chromatin and packing is a Nucleosome (the beads) consisting of ~ 200bp of DNA wound around a nucleosome core particle plus some adjacent linker DNA.
The length of the DNA extracted from the nucleosomes varies between organisms, ranging from 170-240 but the variation results entirely from the linker DNA length between the nucleosomes.
The length of DNA that is wrapped around the histone proteins that make up the core is always about 147 bp base pairs long.
The identity of the proteins that make up the core of the nucleosome came next and included familiar techniques of separating proteins and running them on SDS-PAGE gels.
Revealing that the nucleosome core is an octameric protein complex (two copies each of H2A, H2B, H3, and H4) with the 147 bp DNA wound around it.
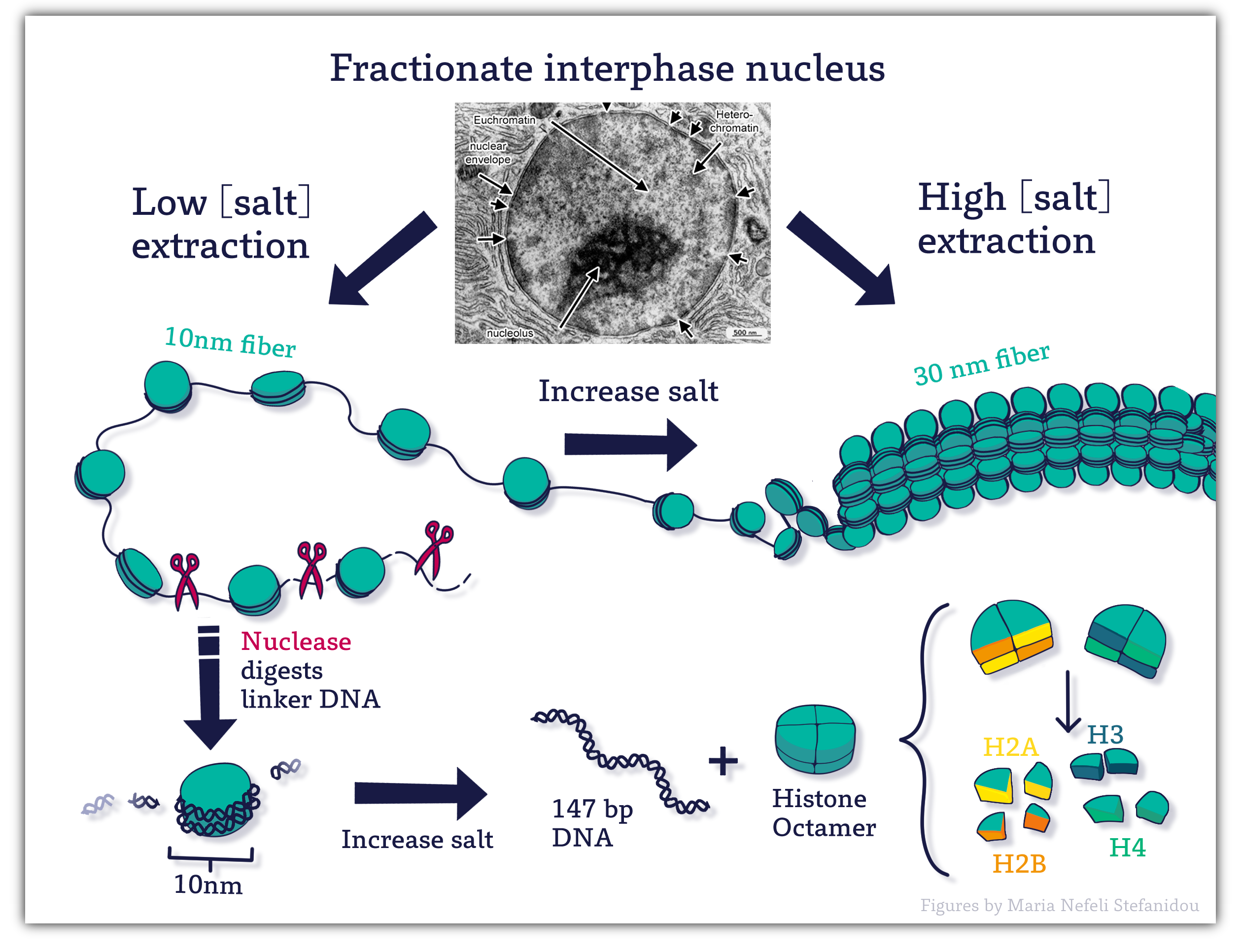 Figure 4.2 Nucleosomes contain DNA wrapped around a protein core of eight histone molecules. Image Credit: Figures by Maria Nefeli Stefanidou (CC-BY-SA-ND)
Figure 4.2 Nucleosomes contain DNA wrapped around a protein core of eight histone molecules. Image Credit: Figures by Maria Nefeli Stefanidou (CC-BY-SA-ND)
Assembly of the Nucleosome
The core histones come together in specific pairs in a very precise arrangement. This association will occur as soon as DNA is replicated!
The assembly of the nucleosome begins with first H3 and H4 forming dimers, the dimers then join to form a tetramer. The DNA begins to wrap around the tetramer and is then joined by the H2A. H2B dimers which cap the complex at the top and bottom (Figure 4. 3) to form the completed nucleosome core particle.
All core histones share a conserved structure known as the histone fold, which consists of three alpha-helices connected by 2 short loops which they use to form the associations within the complex. Importantly, each of the core histone subunits has a short “tail” that sticks out and remains accessible even when the DNA is bound.
This tail is a key feature of the histone, as it is an important site for modification and regulation of the histones as well as of chromatin structure more generally. We will see how these tails influence function in a moment.
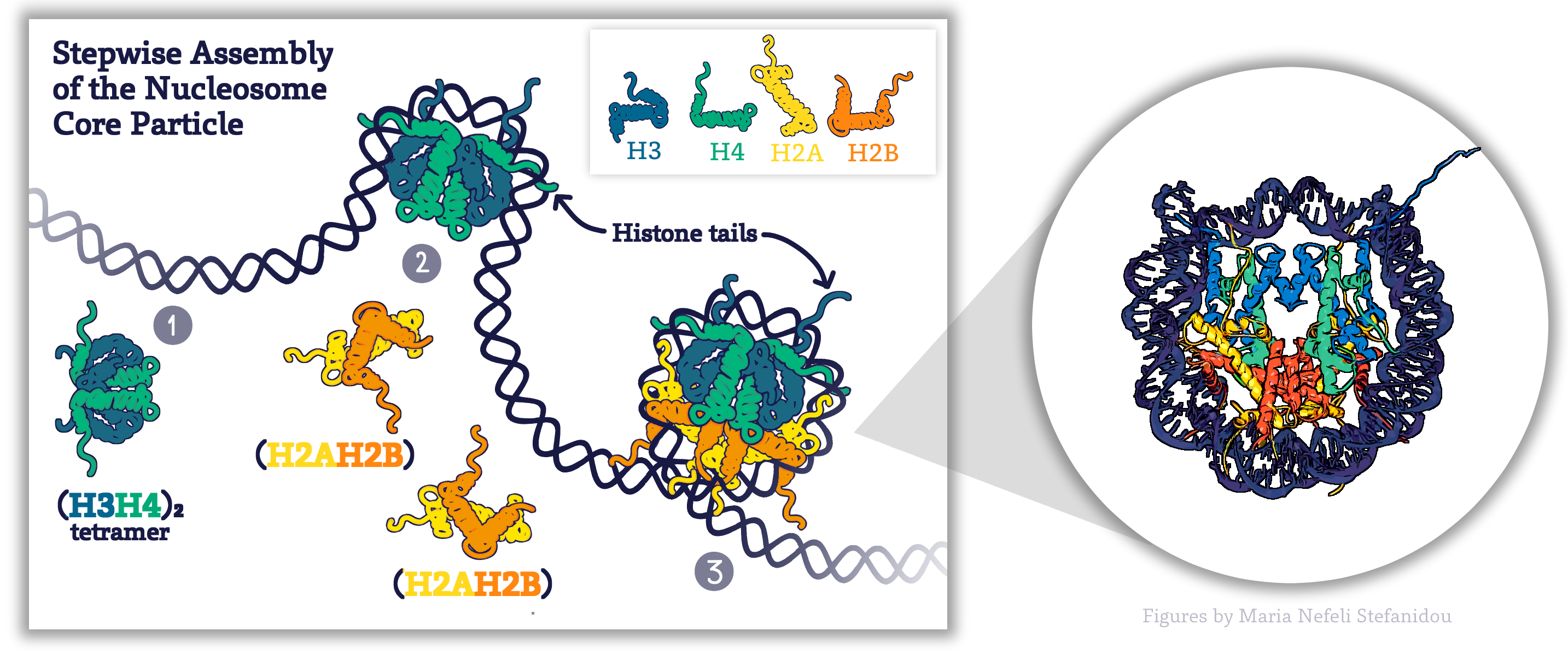
Figure 4.3. Schematic Representation of Stepwise Assembly of the Nucleosome Core Particle, Figure by Maria Nefeli Stefanidou (CC-BY-SA-ND). The structure of nucleosome particles is based on Published NCP Structures (Luger, 2003). Structure Volume 12 Issue 12 Pages 2098-2100 (December 2004) DOI: 10.1016/j.str.2004.11.004
Did I Get This?
Before you continue you should
- Watch the Lecture videos that cover the material above. (if you haven’t already)
Histone-DNA interactions:
The three-dimensional structure of the nucleosome using X-ray crystallography was solved by Dr. Karolin Luger which provided a deeper understanding of chromatin organization.
- Electrostatic interactions of negatively charged DNA (phosphodiester backbone) with positively charged amino acids of histones.
- Additional interactions occur via hydrogen bonds with bases of the DNA.
This association is consistent with the need to package any type of DNA without regard to sequence.
However, there are areas of the genome where nucleosomes position themselves preferentially. These include regions with A-T-rich sequences that are more accommodating of the ‘bend’ or ‘compression’ that occurs in the minor groove as the DNA wraps around the histone octamer.
4.3.3 Second Level of Packing- 30-nm Chromatin Fibres
All chromosomal DNA starts as the “beads-on-string” structure or 10 nm fiber, however, in a living cell, this extended form of structure is not seen. Nucleosomes are further coiled into a shorter, thicker fiber, termed the “30-nanometer chromatin fiber,” (due to average diameter) and the form of DNA that is found in nucleus throughout interphase.
Experimentally this structure can be observed after a high salt chromatin extraction (as shown in Figure 4.2)
The 10nm to 30nm step is believed to be aided by a fifth Histone H1, also known as ‘Linker histone’. In experiments when H1 is present the 30-nanometer fibers form readily.
The role of H1 is different from that of the other histones. H1 does not form part of the nucleosome core, but rather it sits on the surface of the nucleosome, on top of the DNA, and helps keep it in place. It also helps pull in the linker DNA so that the chromatin is more tightly packed.
The tails of the core histones have also been shown to play a role in this second level of packing.
An active interphase genome will naturally show variation in how the genome is packed in different regions—some sections will be packed away tightly (like structural components and genes that are not currently being expressed), and other sections will be more open so that gene expression can take place. This means that even though we discuss this chromatin fiber as if it is always the same, for the purposes of explaining how packing works, we must also remember that the levels of DNA packing are a little more nuanced.
We will explain more when we discuss euchromatin and heterochromatin.
4.3.4 Higher Order Packing
Even though interphase chromatin is well packed compared to the original DNA strand, this is not the end of it. In interphase, when the genome is active, additional packing of the genome must be done in such a way that the genes present on the DNA are taken into account. This is required so that each gene can be easily accessed when needed.
Further during mitosis (and meiosis, which we do not discuss in this textbook), we see the most extreme levels of DNA packing. Each of the chromosomes of the cell must condense itself into the tightest conformation possible and then have its sister chromatids separated into two newly forming daughter cells. Mitotic chromosomes are between 20,000 and 50,000 times shorter than the original DNA strand. No gene expression can happen during this time, which puts the cell at risk, so mitosis is completed as quickly and efficiently as possible.
Thus the 30nm fibers is often folded into 300 nm loops and eventually more compaction into the mitotic chromosome. (Figure 4.4)
Non-Histone Proteins
Non-histone proteins like condensins, scaffold proteins, and cohesins play a role in further coiling and compacting the DNA until it resembles the metaphase chromosome.
Remember that the final metaphase chromosome shape is established and made only in preparation for metaphase. The bulk of the time the cell is in interphase and chromosomes do not resemble those discrete units we are used to thinking of when we think of chromosomes!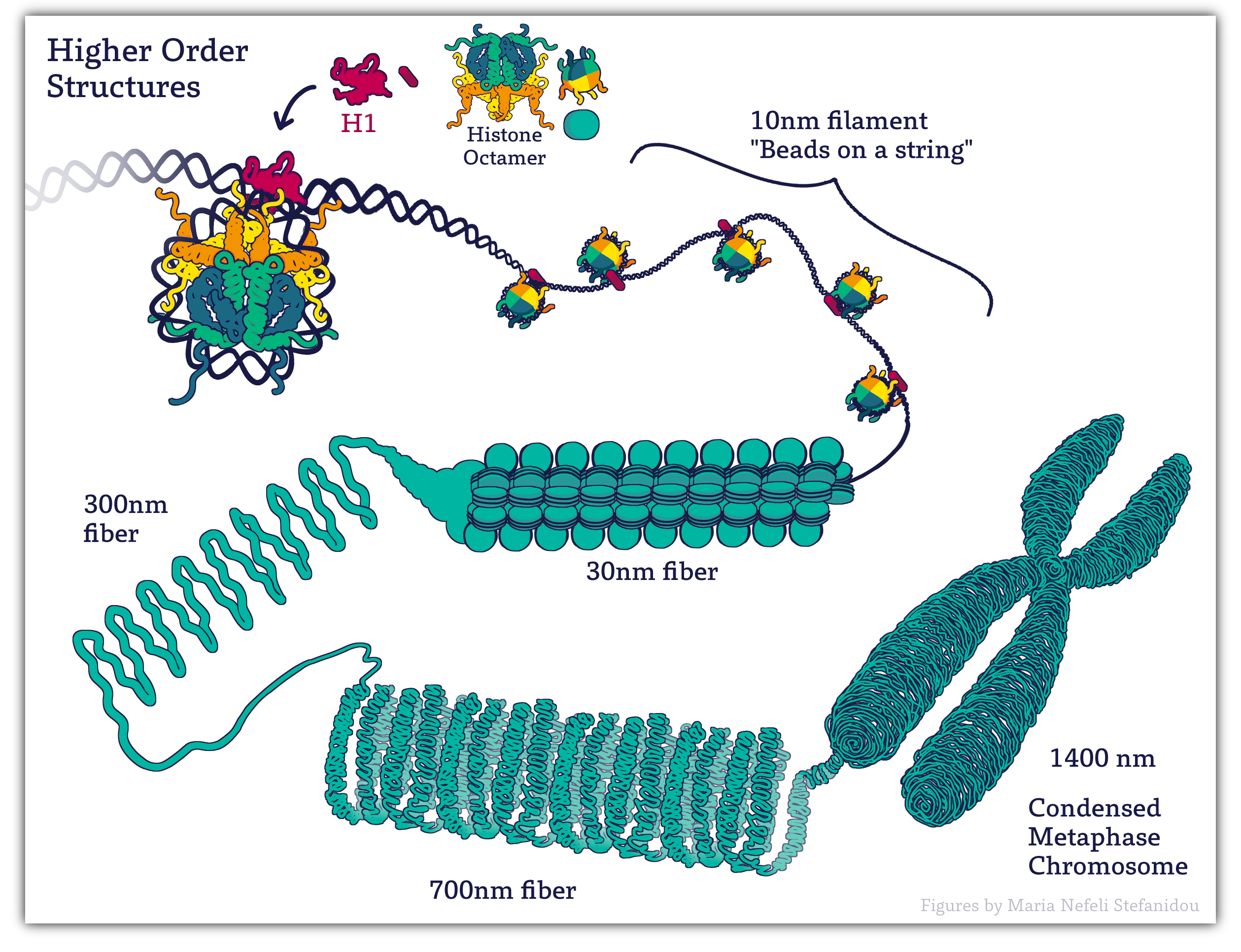
Figure 4.4. Higher orders of chromatin. Image created for this book by Maria Nefeli Stefanidou (CC-BY-SA-ND)
4.3.5 Euchromatin and Heterochromatin
We can now return to forms of chromatin you were introduced to earlier in the chapter. As we have seen mechanisms to pack DNA and maximize space in the nucleus includes ensuring that only the NDA currently needed is unpacked just enough for gene expression. This is the basis for various forms of chromatin seen in interphase nucleus.
The terminology and discovery comes from how they looked under an electron microscope, when they were first observed. The darkly stained material is the heterochromatin, and the lightly stained material is euchromatin.
Heterochromatin is the more tightly packed of the two forms of chromatin. It is the form we described earlier that has the H1 histone bound to it so that the nucleosomes form a spiral and pack together tightly. Heterochromatin does not allow proteins like transcription factors or polymerases to access the DNA. As a result, in these regions no gene expression can take place. At any given moment, most of the chromatin in a cell is in the form of heterochromatin. However, we differentiate between different “types” of heterochromatin:
- Constitutive heterochromatin is found in regions of the DNA that are structural, such as telomeres and/or centromeres. These regions of the DNA never really need to be unpacked, as there are no genes there. Thus, the chromatin stays tightly packed up all of the time so that it takes up less space.
- Facultative heterochromatin, on the other hand, is found in parts of the genome where genes do exist, but they are not currently needed by the cell. These parts are also tightly packed, but if the cell requires one of the genes in this region, it will unpack the DNA to allow for transcription to take place. Thus, these regions may be more dynamic, packing and unpacking as required.
Euchromatin is the less-condensed form of chromatin. These regions are more or less the 10nm or even have all histones removed so that the DNA can be accessed. Active transcription is very likely taking place in these regions as well as other forms of gene regulation. When the genes in these regions are no longer required, they will be packed back up into facultative heterochromatin until they are needed next. These regions of the DNA are considered to be very active and dynamic.
There is one more thing to note before we move on. While all cells will have regions of euchromatin and heterochromatin within the nucleus, the placement of those regions is not always the same from one cell to another. In any given cell, at any given moment, it will be expressing a specific subset of genes. The subset of genes being expressed may or may not be the same as a different cell. This is especially true of cells in different tissue types. A cell of the pancreas synthesizing and secreting digestive enzymes will be expressing a very different set of genes than a neuronal cell, for example. In addition, cells will change the genes that they need to express over time. There are a number of genes that are only turned on during embryonic development and then get turned off. There are a number of genes that are only turned on during embryonic development and then get turned off. Mitosis also requires a specific set of genes that must be expressed to prepare for mitosis and then be turned off again.
All of this has the potential to result in shifts in the parts of the genome that are more or less accessible, which, in turn, will change the regions that are packed as euchromatin or as facultative heterochromatin (since structural regions don’t have genes, constitutive heterochromatin is less likely to change from cell to cell).
How these dynamic changes occurs is the focus of the next section of this chapter.
STUDY TIP: Pause here and watch Dr. Mehta L211 Lecture Video 3: 30nm Fibre and Higher Order Structures
Concepts in Context: Shape of the Genome
Watch:
In this video, you learn how new tools have allowed scientists to probe chromatin organization by finding points of contact between different regions.
COMPLETE: Don’t forget to complete the assignments associated with Concepts in Context within CANVAS.
Key Takeaways
- The primary structure of chromatin is a 10 nm fiber “beads on a string’ structure
- The ‘bead’ is a nucleosome, which includes about 200 bp of DNA wrapped around a histone core consisting of 2 copies each of core histones (H2A, H2B, H3, and H4)
- The DNA closest to the Histone core-core DNA is 147 bp.
- The next level of chromatin is 30 nm fibers- formed by interactions between neighboring nucleosomes.
- Histone H1 is associated with linker DNA and assists in the formation of 30 nm fibers.
- 30nm fibers are folded into higher-order structures (loops upon a proteinaceous scaffold, and then loops are further condensed to form metaphase chromosomes)
- Non-histone proteins are used for higher-order structures.
4.4 Regulation of Chromatin Structure
Watch the Lecture Video first. The section below only provides a summary of the key points from the lecture videos. Click below to take you to the video.
Dr. Mehta Chapter 4 Video 4: Regulation of Chromatin Structure
To recap we have just seen that chromosomal DNA associates with histones, forming an organized complex known as chromatin. This accomplished the goal of allowing DNA to fit into a smaller volume within a eukaryotic cell’s nucleus.
However, compaction of the DNA also limits the accessibility of the DNA to proteins (transcription factors- which we will learn about soon) that will transcribe the gene [Gene Expression].
For gene expression to occur, the chromatin structure needs to move back and forth between condensed and decondensed forms.
Changes to chromatin structure can be brought about in 2 ways and they are not mutually exclusive adding layers of complexity to regulation!
- Via Histone modifications
- Via Chromatin remodeling complexes.
4.4.1 Histone Modifications
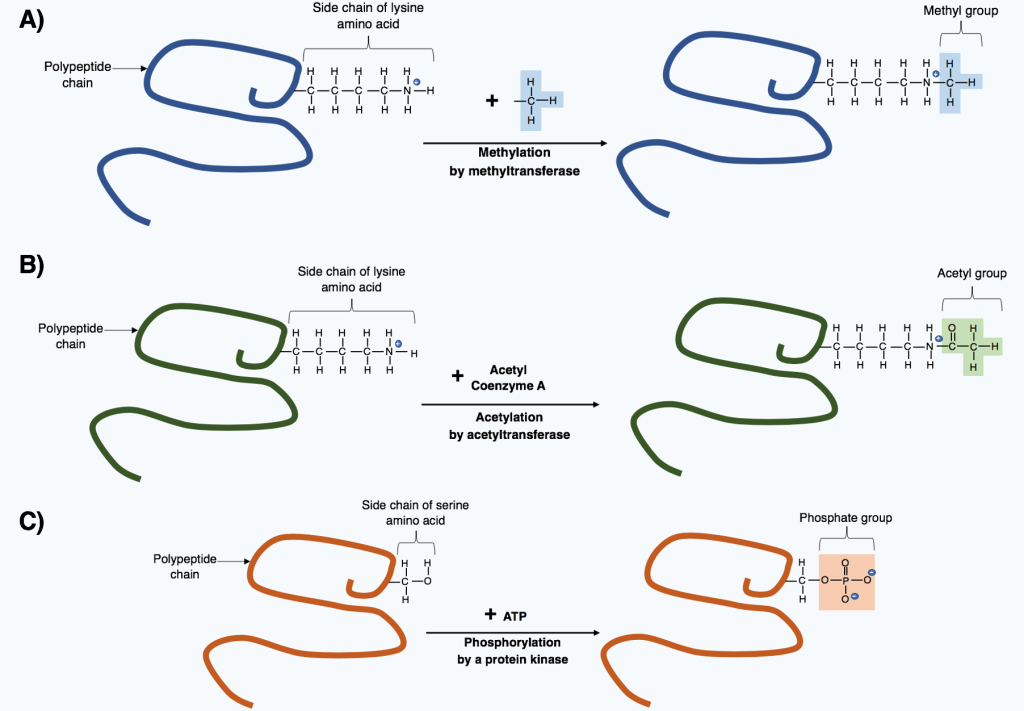
The amino acid residues of histone tails are subject to many post-translational modifications and the list is ever-growing. In particular, histones contain a large percentage of lysine and arginine (basic amino acids) residues and serines, and threonines.
The side chains of these residues are modified by the action of enzymes.
The most studied modifications include acetylation (adding acetyl groups) of lysines, phosphorylation (adding phosphate groups) of serines, and methylation (adding methyl groups) of lysines and arginines.
Connecting Concepts
The addition of the groups is facilitated by enzymes (collectively known as epigenetic “writers”) whilst de-modifying enzymes (or “erasers”) remove these marks. (see table below)
| Histone Acetyl transferases (HATs) | Adds acetyl groups to histones |
| Histone Deacetylases (HDACS) | Removes acetyl groups from histones |
| Histone Methyltrasnderases | Adds methyl groups |
| Histone Demethylases | Remove methyl groups |
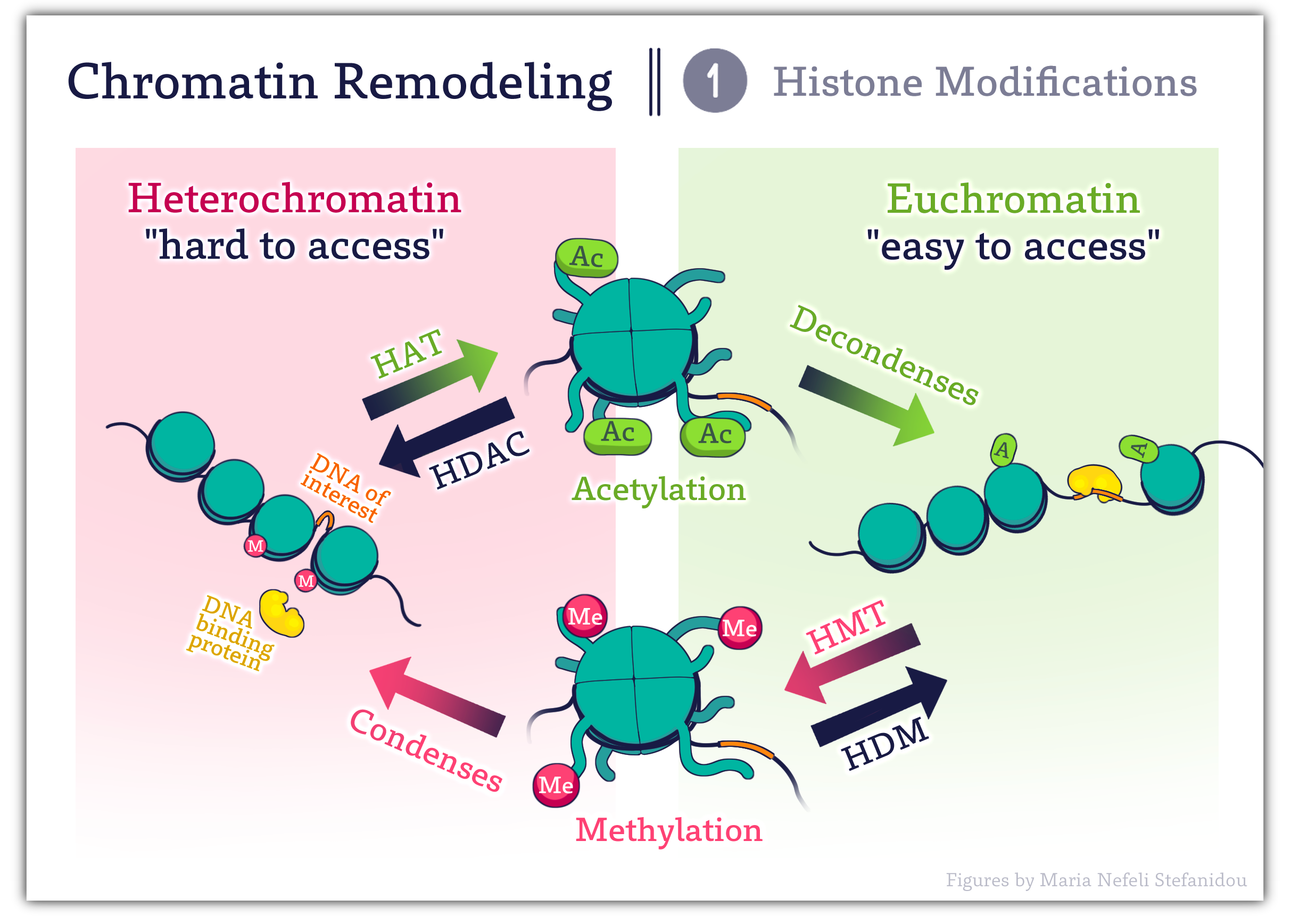
Figure 4.6 Enzymatic modifications of key amino acids within histone tails regulate chromatin structure. Image Credit: Image created for this book by Maria Nefeli Stefanidou (CC-BY-SA-ND)
These opposing activities enable a highly dynamic regulation of gene expression as modifications can be added or removed depending on whether a particular writer or eraser is recruited to a specific location of the genome.
Modifications can occur at different amino acids on different histones and can create more than 100 unique potential changes in the histones.
Modifications also result in unique patterns or signatures that DNA-binding proteins recognize.
Proteins with BROMODOMAINS- recognize Acetylated Lysine residues.
Proteins with CHROMODOMAINS- recognize methylated residues.
To add to the complexity, these proteins and modifying enzymes are contained within a larger multiprotein complex and can recruit another ‘molecular machine’ the chromatin remodeling complexes. These complexes serve to further reposition nucleosomes as discussed below.
4.4.2 Chromatin Remodeling Complexes
- As the name suggests, these are protein complexes meaning they contain multiple different proteins working together.
- They ‘remodel’ chromatin- and alter chromatin structure by repositioning nucleosomes. The mechanisms include
- evicting nucleosome components (such as H2A–H2B dimers)
- ejecting full nucleosomes (creating nucleosome-free regions)
- replacing with variant histone subunits.
There are many families of chromatin remodeling complexes with a variety of different names, and while they are diverse in protein composition they all have at least one enzymatic ‘ATPase subunit which allows them to utilize the energy released from ATP hydrolysis to reposition nucleosomes. (1)
Additionally, as mentioned above they are often recruited to the chromatin by ‘reading’ the ‘marks’ left by histone modifiers OR contain proteins with histone-modifying activities and histone recognition activities.
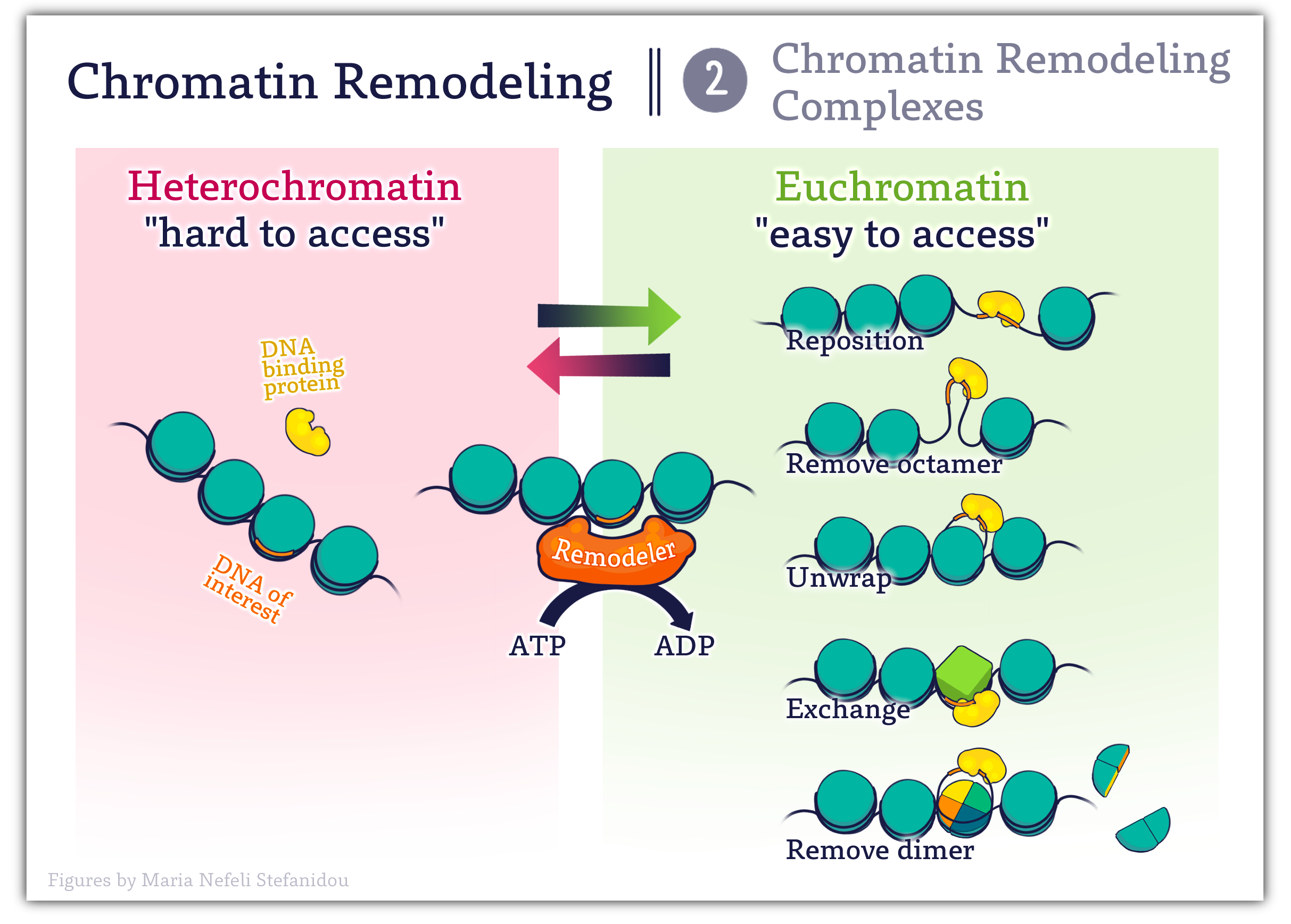
Figure 4.7 Modes of Action of Chromatin Remodeling Complexes. Image created for this book by Maria Nefeli Stefanidou (CC-BY-SA-ND)
4.5 Links to Medicine
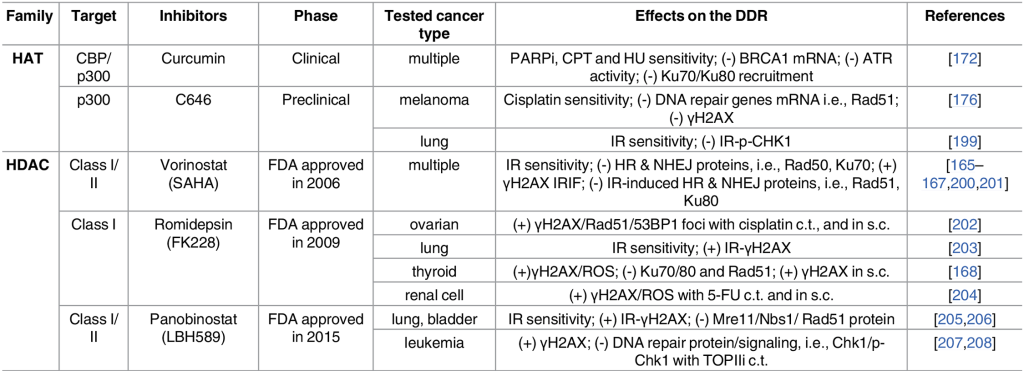
Given the intimate role chromatin structure plays in regulating gene expression, it is not surprising that changes in acetylation signaling resulting from misregulated HATs or HDACs can cause abnormal gene expression patterns and have been identified in numerous cancers. (2)
Some examples are
- Genetic alterations in HATs in hematological and solid cancers (3)
- Hyper-active HDACs in many cancers (4)
- Components of several chromatin-remodeling complexes are highly mutated in cancer.
Not surprisingly there are a wide variety of small-molecule inhibitors targeting acetylation signaling pathways in development for use as anti-cancer drugs (see table below).
Exercises
Remember to:
- Watch the Lecture videos that cover the material above. This will help to clarify or reinforce certain concepts if they are unclear.
- Complete any associated practice exercises
References and Attributions
This chapter contains material taken from the following CC-licensed content and Public Domain content. Changes include rewording, removing paragraphs and replacing them with original material, and combining material.
References
(1) Clapier, C., Iwasa, J., Cairns, B. et al. Mechanisms of action and regulation of ATP-dependent chromatin-remodeling complexes. Nat Rev Mol Cell Biol 18, 407–422 (2017). https://doi.org/10.1038/nrm.2017.26
(2) Gong F, Chiu LY, Miller KM (2016) Acetylation Reader Proteins: Linking Acetylation Signaling to Genome Maintenance and Cancer. PLOS Genetics 12(9): e1006272. https://doi.org/10.1371/journal.pgen.1006272
(3) Di Cerbo V, Schneider R. Cancers with wrong HATs: the impact of acetylation. Briefings in functional genomics. 2013;12(3):231–43. pmid:23325510
(4) Gui CY, Ngo L, Xu WS, Richon VM, Marks PA. Histone deacetylase (HDAC) inhibitor activation of p21WAF1 involves changes in promoter-associated proteins, including HDAC1. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(5):1241–6. pmid:14734806
(5) Terminology Quote from Fundamentals of Cell Biology Copyright © 2024 by Lauren Dalton and Robin Young is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License/

