Molecular Biology: From DNA to RNA to Protein
10 Genetic Code and Translation
10.1 Overview of Translation
Within this chapter, we will cover the details of prokaryotic and eukaryotic translation. Translation is the process of converting the information housed in mRNA into the protein sequence.
Amino acids are linearly strung together via covalent bonds (called peptide bonds) between amino and carboxyl termini of adjacent amino acids. The sequential polymerization of amino acids, in a strict order determined by the sequence of an mRNA, is catalyzed by a ribonucleoprotein complex called the ribosome working with decoding “keys” termed charged tRNAs.
Recall that prokaryotic and eukaryotic transcription and translation systems differ in large part due to the compartmentalization of larger eukaryotic cells. Due to this compartmentalization, transcription and translation are separated spatially and temporally within the cell. Transcription occurs within the nucleus of eukaryotes and translation occurs within the cytoplasm (Fig. 10.1 B). Prokaryotes do not have compartmentalization and have, thus, evolved a coupled transcription/translation system where both process occur simultaneously (Fig. 10.1 A).

Figure 10.1 Cellular Location of Transcription and Translation in Prokayotes and Eukaryotes. (a) Prokaryotes lack cellular compartmentalization and show coupled transcription-translation processing, whereas (b) eukaryotes have a high degree of compartmentalization and separate the processes of transcription, which is in the nucleus of the cell, from the processes of translation, which is localized in the cytoplasm.Figure from: Baccei, A., and Rice, M. Lumen Learning
Recall that peptide formation is a dehydration reaction that combines the carboxylic acid of the upstream amino acid with the amine functional group of the downstream amino acid to form an amide linkage (Fig. 10.2). Water is the by-product. The ribosome (a large complex of peptides and rRNA molecules) serves as the enzyme that mediates this reaction. It requires a mature mRNA to serve as the template, and performs peptide bond synthesis in a directional fashion from the N to the C-terminal of the growing peptide/protein.
This is known as N- to C-synthesis.
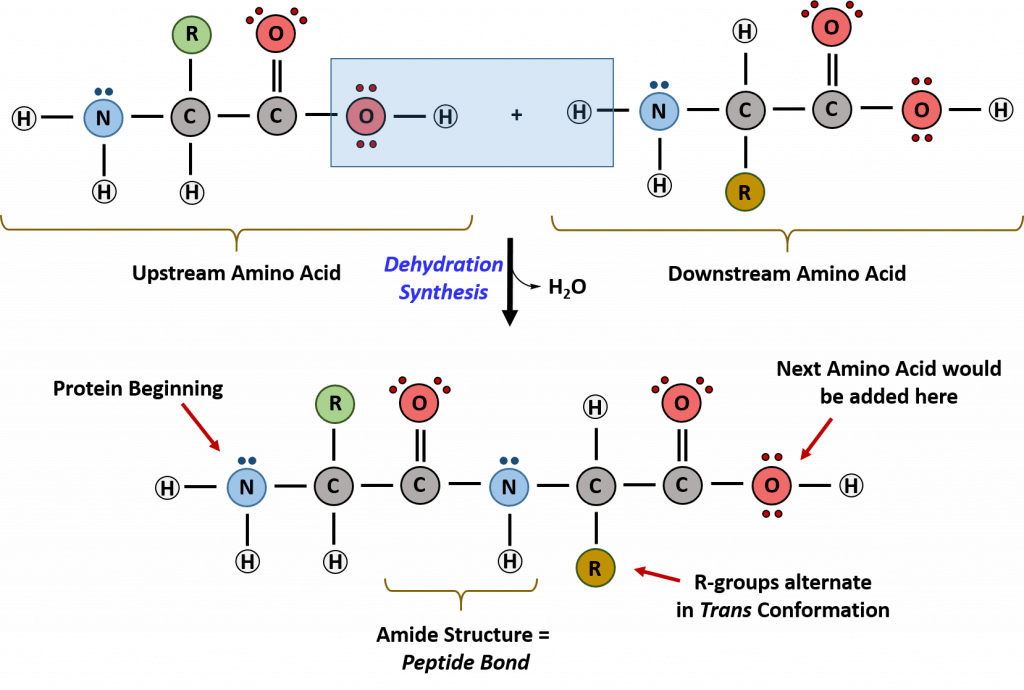
Figure 10.2 Formation of the Peptide Bond. The addition of two amino acids to form a peptide requires dehydration synthesis. The carboxylic acid of the upstream amino acid is joined with the amine functional group of the downstream amino acid to form the amide linkage. Within the ribosome, this reaction is highly directional and only occurs in the N to C orientation. Figure from: Flatt, P.M. (2019) Biochemistry – Defining Life at the Molecular Level. Published by Western Oregon University, Monmouth, OR (CC BY-NC-SA). Available at: https://wou.edu/chemistry/courses/online-chemistry-textbooks/ch450-and-ch451-biochemistry-defining-life-at-the-molecular-level/chapter-11-translation/
Learning Objectives
- Explain what the terms nonoverlapping, commaless (without punctuation), degenerate, and unambiguous mean with respect to the genetic code.
- Define the following terms as they apply to the genetic code: Reading frame, Initiation codon , Termination codon , Sense Codon
- What is the wobble hypothesis and how does it fit with the fact that the genetic code is degenerate?
- Be familiar with the genetic code and be able to use it to deduce the primary structure of a polypeptide from an mRNA sequence
10.1.2 Overview of Genetic Code
We speak of genes (i.e., DNA) coding for proteins and the central dogma, which states that DNA makes RNA make protein.
What does this actually mean? A code can be thought of as a system for storing or communicating information.
Analogy: A familiar example is the use of letters to represent the names of airports (e.g., PDX for Portland, Oregon, and ORD for Chicago’s O’Hare). When a tag on your luggage shows IND as the destination, it conveys information that your bag should be sent to Indianapolis, Indiana.
To function well, such a setup must have unique identifiers for each airport and people who can decode the identifiers correctly. That is, IND must stand only for Indianapolis, Indiana, and no other airport. Also, luggage handlers must be able to correctly recognize what IND stands for so that your luggage doesn’t land in Iowa, instead.
How does this relate to genes and the proteins they encode?
Genes are first transcribed into mRNA, as we have already discussed. The sequence of an mRNA, copied from a gene, directly specifies the sequence of amino acids in the protein it encodes.
The genetic code is the information for linking amino acids into polypeptides in an order based on the base sequence of 3-base codewords (codons) in a gene and its messenger RNA (mRNA).
For example, the amino acid tryptophan is encoded by the sequence UGG on an mRNA. All of the twenty amino acids used to build proteins have, likewise, 3-base sequences that encode them.
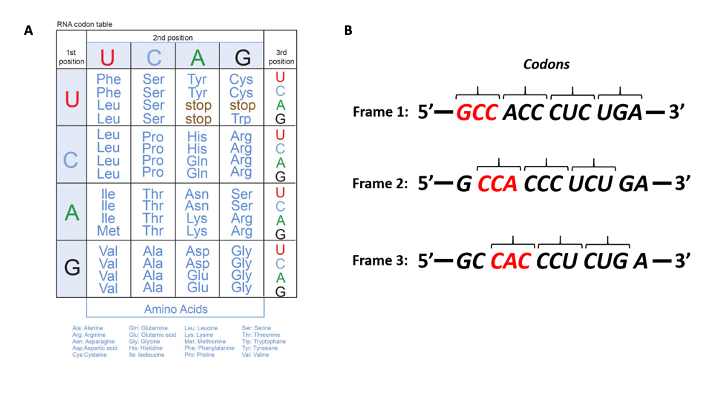
Fig. 10.3A (below) shows representations of the genetic code in the ‘language’ of RNA.
The left-hand vertical column indicates the first (5’) position in a codon, the horizontal bar across the top indicates the second position, and the right-hand vertical column indicates the third (3’) position
10.1.3 Features of the Genetic Code
Three nucleotides encode an amino acid
Template mRNA is read by the ribosome in groups of three nucleotides, called a codon (Fig. 10.3 B). Simple calculations hypothesized ( a minimum of 3 bases would be needed to code for 20 amino acids) and genetic experiments ultimately proved this to be the case.
Non-overlapping and Unambiguous
The template is non-overlapping and reads in discrete groups of three. This is known as the reading frame of the mRNA, and it is always read from the 5′ to 3′ direction.
Thus, for each mRNA, there are three potential reading frames (Fig. 10.3A). Only one reading frame will be the correct one for protein synthesis.
The ribosome must recognize and align the correct reading frame of the mRNA such that the correct codon sequences can be read.
Look at the codon chart in Figure 10.3A. We can see that each codon is specific for a single amino acid.
For example, UUU is always coding for Phenylalanine and not any other amino acid.
There is very little ambiguity within the code.
There is no punctuation
The sequence of bases is read continuously without stopping or skipping nucleotides.
Degeneracy and Redundancy
Given that there are 4 bases in RNA, the number of different 3-base combinations that are possible is 43 or 64. There are, however, only 20 amino acids that are used in building proteins in cells.
This discrepancy in the number of possible codons and the actual number of amino acids they specify is explained by the fact that the same amino acid may be specified by more than one codon.
In fact, with the exception of the amino acids methionine and tryptophan, all the other amino acids are encoded by multiple codons.
Codons for the same amino acid are often related, with the first two bases the same and the third being variable.
An example would be the codons for alanine: GCU, GCA, GCC, and GCG all stand for alanine.
This sort of redundancy in the genetic code is termed degeneracy.
Rules of translation: Stop and start codons
All codons that code for an amino acid are also referred to as SENSE Codons.
Whereas 61 of the 64 possible triplets code for amino acids, three of the 64 codons do not code for an amino acid; they terminate protein synthesis, releasing the polypeptide from the translation machinery.
The three stop codons are UAA, UGA, and UAG. The three-stop codons also have colloquial names: UAA (ochre), UAG (amber), UGA (opal), with UAA being the most common in prokaryotic genes.
In contrast, evolution has selected the codon for methionine, AUG, as the start codon for all polypeptides (regardless of their function) and for the insertion of methionine within a polypeptide.
Thus, all polypeptides begin life with a methionine at their amino-terminal end!
Open Reading Frame
As mentioned above when the genetic code is read on mRNA there are three potential reading frames.
The frame is set by the AUG start codon near the 5’ end of the mRNA. Each set of three nucleotides following this start codon is a codon in the mRNA message until the termination codon is the reading frame.
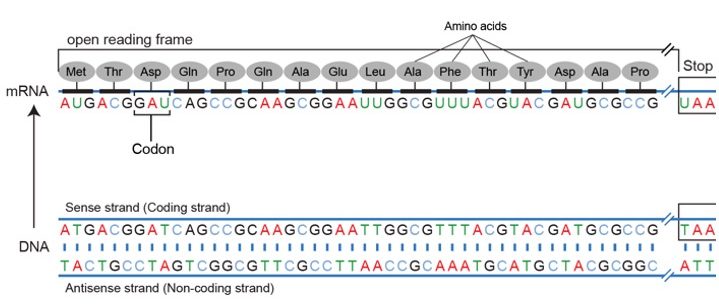
Using the same diagram as above we can see that of the potential reading frames only one of them makes an intelligible protein.
The first terminates immediately, the second runs into the end, and still no termination codon.
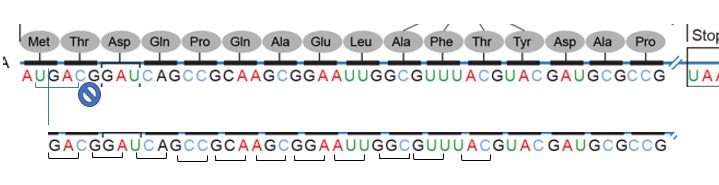
In practice usually, the one with the longest stretch of codons is typically indicative of an open reading frame.
Did I get this? Concept of Open Reading Frames
When scanning a genome for genes that may encode proteins, scientists use bioinformatics programs like ORF Finder to look for start codons, stop codons, and stretches of DNA in between the two that code for proteins at least 50 to 300 amino acids long.
Other clues include the presence of promoter sequences ahead of the start codon.
These open reading frames can then be analyzed further, using bioinformatics tools like BLAST searches and phylogenetic analyses to determine whether these areas are similar to other known genes from
other organisms, which may then warrant further study in the lab.
Gene sequences are largely conserved – so if an ORF sequence is present in multiple genomes, it likely represents a gene!
Q. For a double-stranded DNA molecule how many reading frames are possible?
(Nearly) Universal
With a few exceptions and with minor changes (some prokaryotes, mitochondria, chloroplasts, ciliated protozoa), the genetic code is the same in all organisms from viruses and bacteria to humans, providing support for a single origin of life.
Exceptions include some protozoans using UAA and UAG as codons for amino acids rather than as stop signals; UGA is their sole termination signal. Mitochondrial DNA encodes for a distinct set of mitochondrial tRNAs;s which can recognize alternative codons. Thus the genetic code is nearly universal.
Practice: How to Read a Codon Chart
Molecular Biology in the News: Concepts in Context
Read the associated article (PDF files on CANVAS if the link above is pay walled).
As you read think about:
The criteria that scientists used for identifying genes and why these mini proteins were missed and how the method to determine which proteins cells are making that as developed helped them find these proteins.
Learning Objectives
- What is the adaptor hypothesis and how does the tRNA fit into this hypothesis?
- Be able to predict how mutations in the anticodon section of a tRNA will affect the ‘translation’ of mRNA.
- Describe the roles and relationships between tRNA synthetases and tRNA molecules, tRNA anticodon sequences, and mRNA codon sequences. How are tRNAs linked to their corresponding amino acids?
- Bacteria have two different kinds of tRNA. What roles do these tRNAs play in polypeptide synthesis?
-
How does translation initiation in eukaryotes differ from that in prokaryotes?
- Describe/illustrate or be able to label the initiation stage in bacterial polypeptide synthesis. Discuss the role of each protein factor that participates in this process.
- Give the elongation factors used in bacterial translation and explain the role played by each factor in translation.
-
What is the role of codons UAA, UGA, and UAG in translation? What events occur when one of these codons appears at the A site of the ribosome?
- Compare and contrast the process of protein synthesis in bacterial and eukaryotic cells, giving similarities and differences in the process of translation in these two types of cells.
- In a diagram of translation be able to label:
5′ and 3′ ends of the mRNA; A, P, and E sites; Start codon; Stop codon; Amino and carboxyl ends of the newly synthesized polypeptide chain; Approximate location of the next peptide bond that will be formed; Place on the ribosome where release factor 1 will bind
LEVEL UP
- Predict the effect of a mutation in the part of the tRNA gene that changes: (a) the acceptor stem; (b) the anticodon
- In a cell-free protein-synthesizing system (a test tube) predict the effects of omitting initiation or elongation factors in translation. ( What, if any, type of protein would be produced? Explain your reasoning.)
- Predict the effect or stage of translation at which antibiotic affects protein synthesis when given a scenario or description.
10.2 tRNA’s: The Interpreter of the Code
While the ribosomes are the factories that join amino acids together using the instructions in mRNAs, another class of RNA molecules, the transfer RNAs (tRNAs) are also needed for translation.
In terms of the bead analogy above, someone or something has to be able to bring a red bead in when the instructions indicate UGG, and a green bead when the instructions say UUU. This, then, is the function of the tRNAs.
They act as ADAPTORS or interpreters of the code. They act as adaptors by binding to the codon on one end and carrying the amino acid on the other end.
There is at least one tRNA for each amino acid.
All transfer RNAs share common features and are structurally similar. These include
1. Transfer RNAs are small single-stranded RNA molecules, about 75-90 nucleotides long.
2. Transfer RNAs are extensively modified post-transcriptionally and contain a large number of unusual bases and modified bases like Inosine.
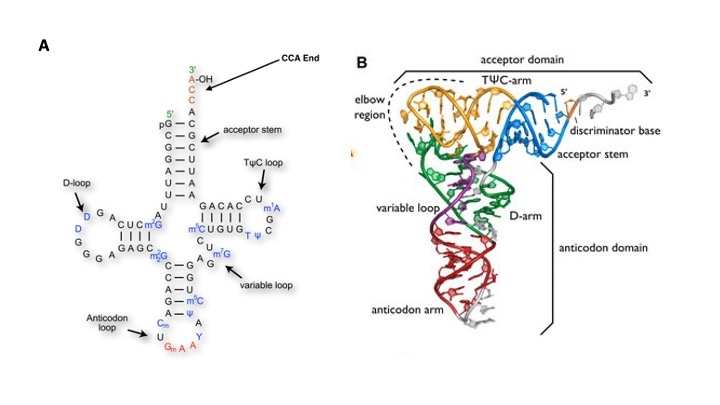
3. Mature tRNAs take on a three-dimensional structure where the single-stranded tRNA folds on itself and base-pairs to form what is sometimes described as a stem-loop, or cloverleaf structure.
This structure is crucial to the function of the tRNA, providing both the sites for attachment of the appropriate amino acid and for recognition of codons in the mRNA.
The cloverleaf consists of five parts: the acceptor stem (containing the tRNA’s 5′- and 3′-ends), the D-arm, the anticodon arm, the variable loop, and the TΨC-arm (T-arm).
4. All tRNAs contain at the 3′-terminus (acceptor stem) the nucleotides CCA. These are added to the tRNA post-transcriptionally by CCA-adding enzymes.
5. For all tRNA amino acid is attached to a hydroxyl group of the A (of the CCA sequence).
6. At the other end of the acceptor’s arm is the anticodon loop.
Every tRNA has a sequence of 3 bases, the anticodon, that is complementary to the codon for the amino acid it is carrying. When the tRNA encounters the codon for its amino acid on the messenger RNA, the anticodon will base-pair with the codon.
Note that the pairing of anticodon with codon within the message like all other forms of nucleic acid interactions is ‘antiparallel’.
Also note that the sequences are both written, by convention, in the 5’ to 3’ direction as we have seen earlier for all written forms.
For the tryptophan tRNA this is what it would look like:
The sequence of tryptophan codon in mRNA: 5’ -UGG- 3’
The codon-anticodon basepair in the antiparallel orientation then would be:
5’ – UGG -3’
3’- ACC- 5’
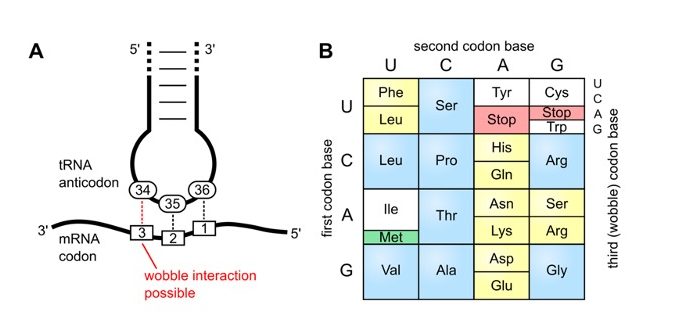
Wobble Base Pairing
The degeneracy of the genetic code – where many tRNA molecules can recognize more than one codon using a single anticodon is due to a feature known as ‘Wobble Base Pairing”.
A wobble base pair is a pairing between two nucleotides in RNA molecules that does not follow Watson-Crick base pair rules.
The four main wobble base pairs are guanine-uracil (G-U), hypoxanthine-uracil (I-U), hypoxanthine-adenine (I-A), and hypoxanthine-cytosine (I-C).
The wobble base position is usually the first position of the anticodon (read in the 5′ – 3′ direction), which aligns with the 3rd position of the mRNA codon.
In order to maintain consistency of nucleic acid nomenclature, “I” is used for hypoxanthine because hypoxanthine is the nucleobase of the inosine nucleotide– one of the modified bases on tRNA’s
The thermodynamic stability of a wobble base pair is comparable to that of a Watson-Crick base pair.
Wobble base pairs are fundamental in RNA secondary structure and are critical for the proper translation of the genetic code.
Did I get this?
A series of tRNAs have the following anticodons. Consider the wobble rules listed in Table 10.6, and give all possible codons with which each tRNA can pair with
A) 5′−GGC−3′
B) 5′−AAG−3′
Key Takeaways
10.2.1 Charging of tRNA’s- Amino Acyl tRNA Synthetases
The fidelity (accuracy) of protein synthesis is maintained by the ribosome’s ability to match the code from the template mRNA strand with the appropriate amino acid.
However, it is the tRNA that forms a physical link between the mRNA and the amino acid the codon represents.
Therefore, the accuracy of translation hinges upon the very important process of ensuring that the correct amino acid is added to the appropriate tRNA!
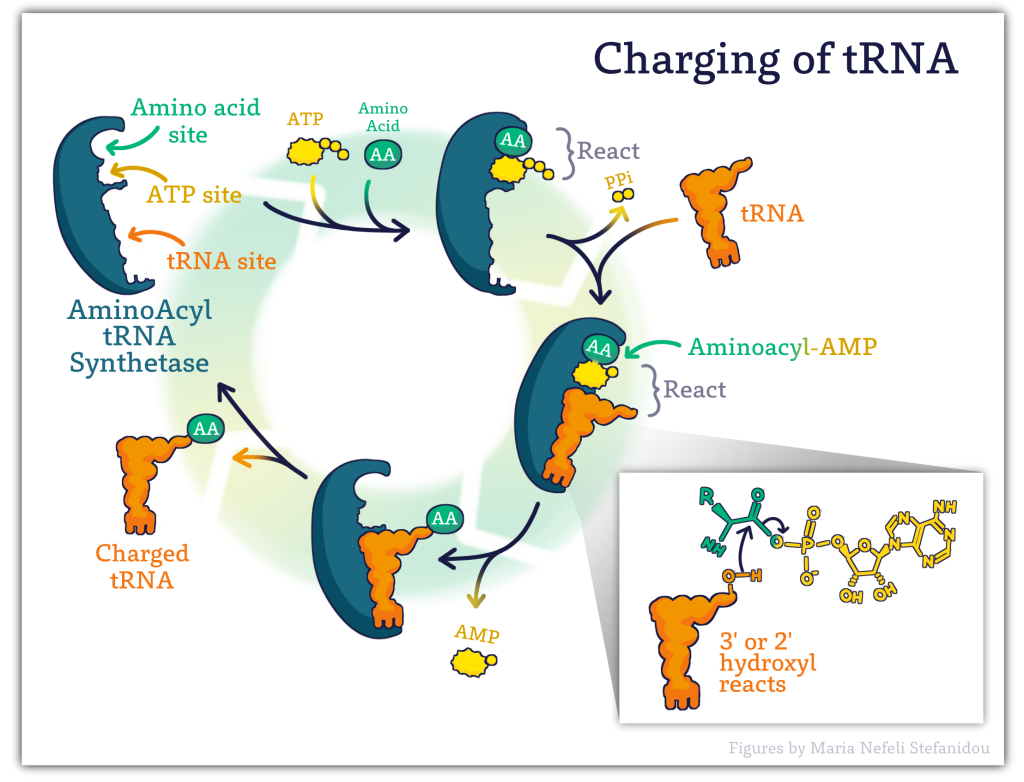
Before the tRNA is brought to the ribosome, amino acids are attached to the tRNA! The attachment of amino acids to the tRNA is known as the ‘Charging of tRNA’. A pool of charged tRNAs is necessary to carry out protein synthesis.
The recognition and attachment of amino acids to tRNAs are carried out by a class of enzymes called: Aminoacyl tRNA synthetases.
There is a different synthetase enzyme for each amino acid, therefore there are 20 aminoacyl-tRNA synthetases- ONE for each of the 20 amino acids!
They are named after the aminoacyl-tRNA product generated, as such, methionyl-tRNA synthetase (abbreviated as MetRS) charges tRNAMet with methionine.
Charged tRNAs are often indicated in written form as with the amino acid in superscript. Example: met-tRNAMet
In order to ensure the faithful translation of the genetic message, synthetases must identify and pair particular tRNAs with their corresponding amino acid which relies on the proper recognition of both substrates- the tRNA and the amino acid. The enzymes thus have developed an elevated specificity for both substrates.
There is also a built-in pre-attachment proofreading mechanism in that tRNA molecules that fit the synthetase well (i.e. the correct ones) maintain contact longer and allow the reaction to proceed whereas ill-fitting and incorrect tRNA molecules are likely to disassociate from the synthetase before it tries to attach the amino acid.
The overall process involves the binding of both the amino acid and specific tRNA on separate binding sites. ATP then binds the enzyme. The final product (Charged tRNA) is formed in 2 steps with the first linking the amino acid to the ATP- in a high energy bond , the amino acid (AA) is then transferred from the AMP to either the 2′-OH of the terminal A (of CCA of tRNA) or 3′-OH of terminal A (of CCA of tRNA) tRNA. See Figure 10.7 below.
Watch Dr. Mehta’s Lecture Video on Aminoacyl tRNA synthetases (6 minutes)
10.3 Ribosome Structure
The ribosome is a highly conserved molecular machine.
In all organisms, it is composed of two unequal subunits, called LARGE and SMALL subunits. Each consists of a distinct set of ribosomal RNA (rRNA) and ribosomal proteins (RPs) that combine to form a large nucleoprotein complex.
Prokaryotic ribosomes: have a mass of about 2500 kDa and a size of 70S (or Svedberg units: A Svedberg unit is a measure of the sedimentation rate in a centrifuge and thus is representative of size).
A complete ribosome (70S) can be dissociated into a large subunit (50S) and a small subunit (30S)
Eukaryotic ribosomes are larger than their prokaryotic counterparts at approximately 80S (although there is some modest variation between eukaryotic species). Human cytosolic ribosomes are composed of a large subunit (60S) and a small subunit (40S).
The ribosome structures in all living organisms regardless of size however function similarly and carry out three important tasks.
1) Bind the mRNA and find the start codon, where the translation will begin
2) Facilitate (provide a place) for the tRNA to come in and ‘decode’. Molecularly- facilitate the complementary base pairing of mRNA codons and tRNA anticodons that determines amino acid order in the polypeptide.
3) Catalyze the peptide bond formation between the amino acids.
Structurally they harbor three different tRNA binding sites that help the ribosomes carry out these tasks.
The A-site, where decoding occurs and the correct aminoacyl-tRNA (aa-tRNA) is selected on the basis of the mRNA codon displayed.
During protein synthesis tRNA’s charged with amino acid are entering here.
The P-site, which holds the peptidyl-tRNA, (the growing polypeptide)
The E-site binds exclusively to deacetylated tRNAs (uncharged tRNAs) that are exiting the ribosome.
Thus, during translation the tRNA moves from the A-site through the P- and E-site, where it leaves the ribosome (Fig. 10. 8).
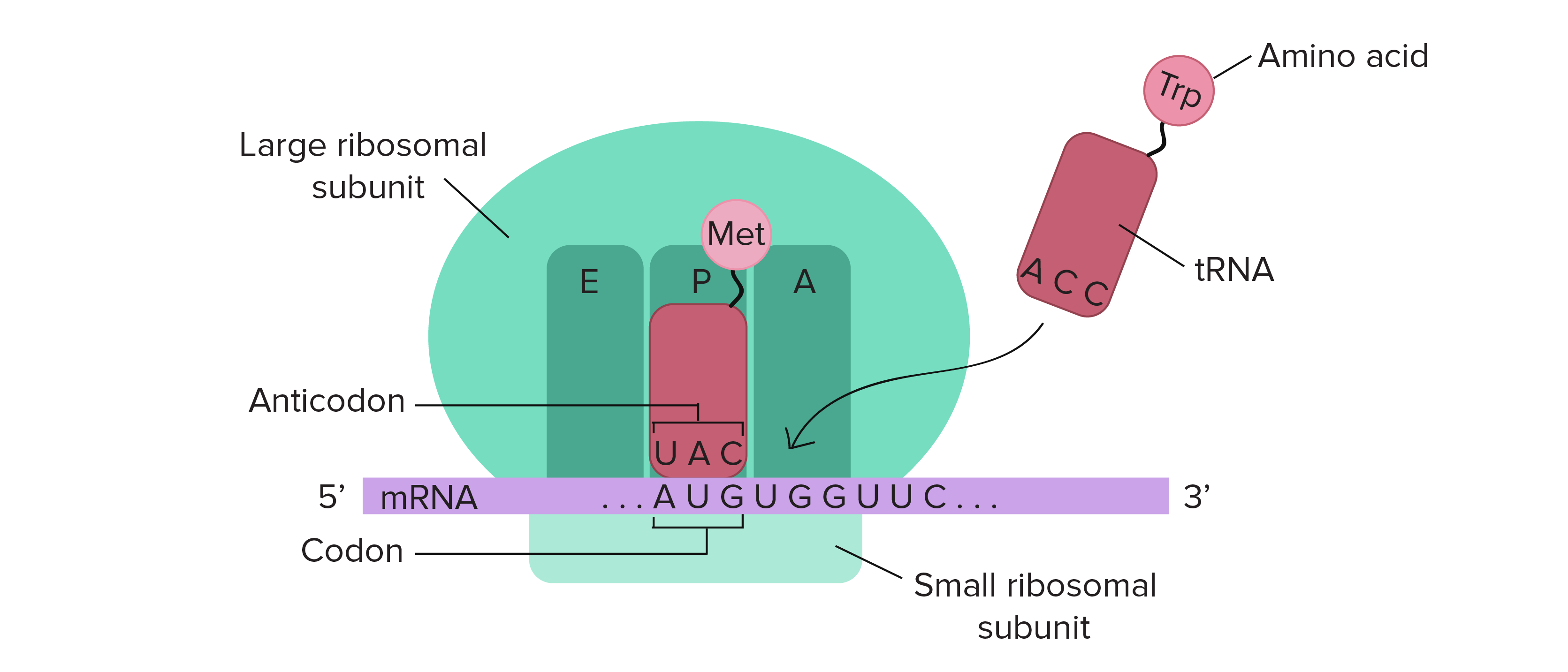
Figure 10.8 Schematic Structure of an Active Ribosome. The mRNA (shown in purple) is assembled between the small subunit and the large subunit of the ribosome (shown in green). tRNA molecules (shown in red) that are loaded with their cognate amino acid (shown in pink) are transitioned through the A-P-E sites of the ribosome during the elongation phase of translation. The movement of the tRNA molecules also shifts the position of the mRNA causing the next three codon bases to line up in the A-site of the ribosome.
Figure from: The Khan Academy where it was modified from Openstax College Biology
10.4 Overall Steps in Translation
Translation occurs in three phases and involves cycling in and of tRNAs through the ribosome sites mentioned above.
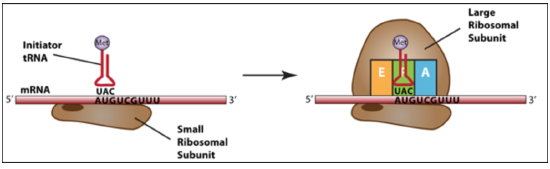
Initiation
Finding the correct AUG sets the open reading frame. (Needs initiator tRNAs and initiation factors).
At the end of initiation, the start codon (AUG) is positioned to base pair with the tRNA in the P-site (peptidyl site).
This is the only time tRNA charged with amino acids occupies the P-site.
Elongation: joining of adjacent amino acids –carried by the tRNA successively.
A tRNA bound to its amino acid (known as an aminoacyl-tRNA) that is able to base pair with the next codon on the mRNA arrives at the A site.
The preceding amino acid (Met at the start of translation) is covalently linked to the incoming amino acid with a peptide bond.
The bond between the amino acid and the tRNA in the P-site is broken and the dipeptide is joined to the tRNA on the A-site.
The initiator tRNA moves to the E site and the ribosome moves one codon downstream. This shifts the most recent tRNA from the A site to the P site, opening up the A site for the arrival of a new aminoacyl-tRNA.
This cycle continues, with the ribosome moving on the mRNA one codon at a time, until the stop codon reaches the A-site.
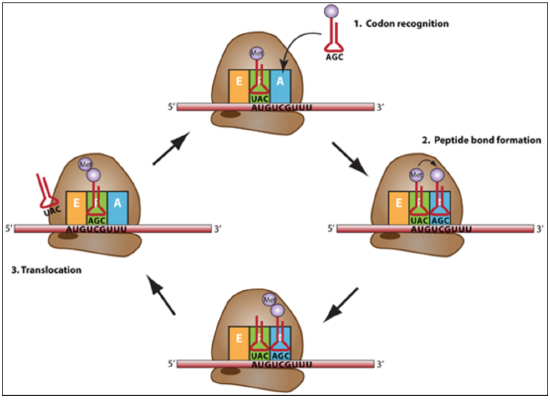
Termination: Termination codons are recognized by release factors. The completed polypeptide chain is released.
The ribosome then dissociates into the small and large subunits, once more.
Link to Learning
Polysomes
Each mRNA molecule is simultaneously translated by many ribosomes, all synthesizing protein in the same direction: reading the mRNA from 5’ to 3’ and synthesizing the polypeptide from the N terminus to the C terminus.
The complete structure containing an mRNA with multiple associated ribosomes is called a polyribosome (or polysome). In bacteria, before transcriptional termination occurs, each protein-encoding transcript is already being used to begin the synthesis of numerous copies of the encoded polypeptide (s) because the processes of transcription and translation can occur concurrently, forming polyribosomes This allows a prokaryotic cell to respond to an environmental signal requiring new proteins very quickly.
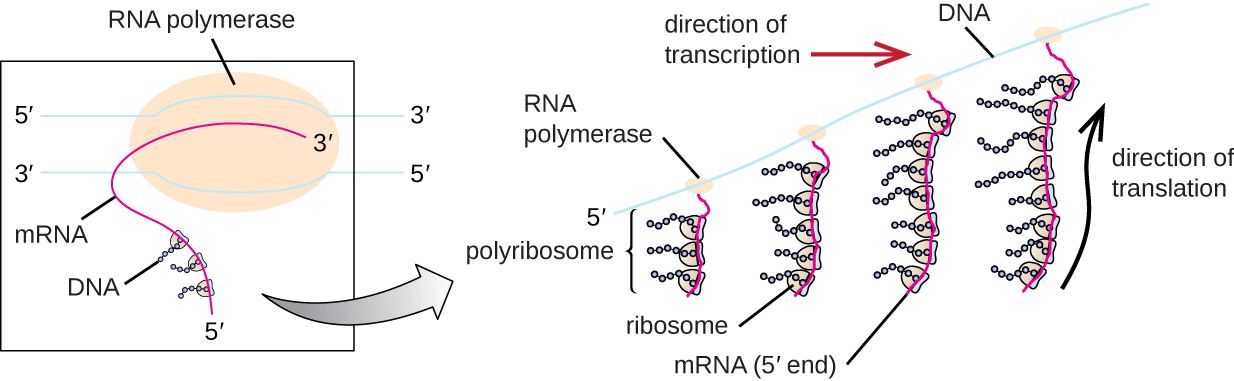
Figure 10.9
In prokaryotes, multiple RNA polymerases can transcribe a single bacterial gene while numerous ribosomes concurrently translate the mRNA transcripts into polypeptides. In this way, a specific protein can rapidly reach a high concentration in the bacterial cell. Figure from: “Protein Synthesis (Translation)” by OpenStax, LibreTexts is licensed under CC BY .
Watch this Animation at https://www.labxchange.org/
Concept Check
Concept: In both prokaryotic and eukaryotic cells, multiple ribosomes may translate a single mRNA molecule simultaneously, generating a structure called a polyribosome.
In a polyribosome, the polypeptides associated with which ribosomes will be the longest?
a) Those at the 5′ end of mRNA
b) Those at the 3′ end of mRNA
c) Those in the middle of mRNA
d) All polypeptides will be the same length.
Answer at end.
10.5 Details of Translation
Having considered the steps of translation in broader terms, we can now look at them in greater detail.
As in all the processes we have learned, the first step Initiation is where most of the differences occur between Prokaryotic and Eukaryotic Translation.
10.5.1 Initiation of Protein Translation
There are three steps to translation initiation (achieved differently in eukaryotes and in bacteria) that conceptually involve
1. Identifying the start codon (involves the small ribosome subunit)
2. Positioning the initiator tRNA in the P-site
3. Forming the active complex by joining of large ribosome subunit.
Protein factors called initiation factors facilitate these steps and ensure speed and accuracy in the overall process.
Messenger RNAs have non-coding sequences both at their 5′ and 3′ ends, with the actual protein-coding region sandwiched in between these untranslated regions (called the 5′ UTR and 3′ UTR, respectively).
The ribosome must be able to recognize the 5′ end of the mRNA and bind to it, then determine where the start codon is located.
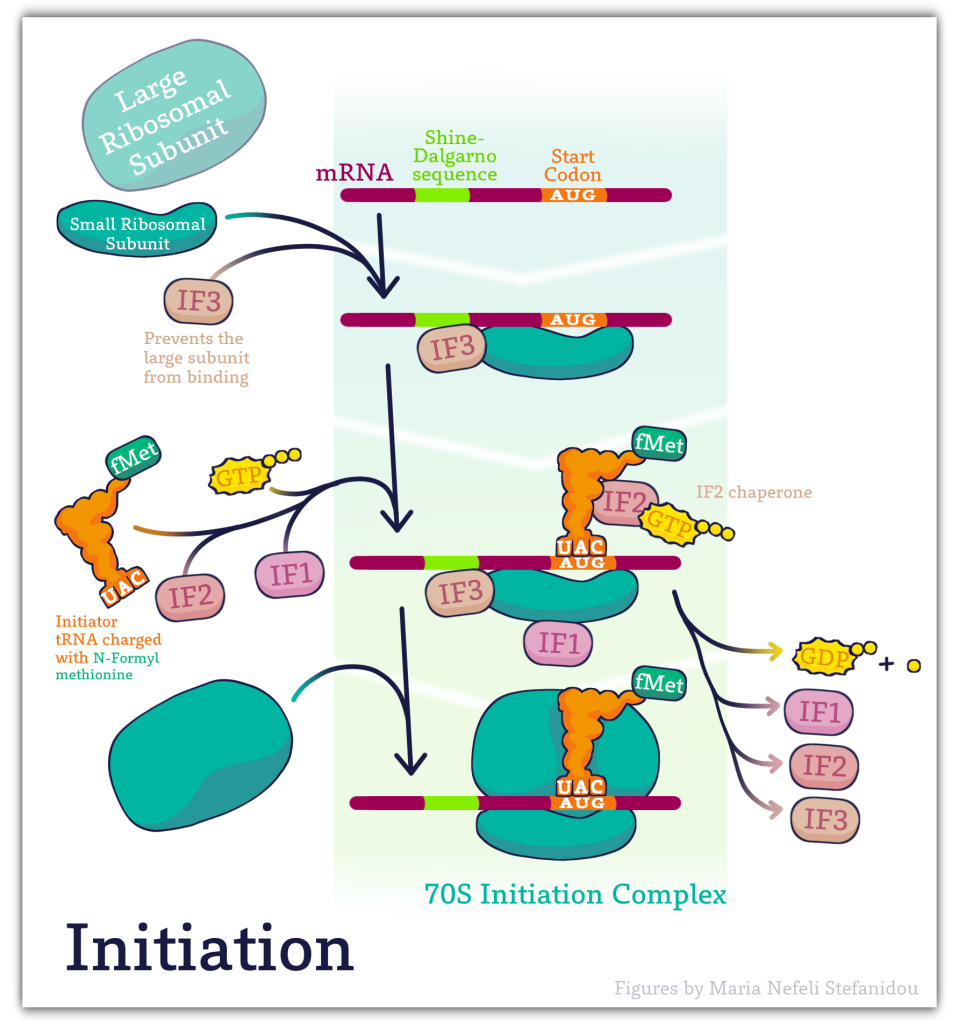
10.5.2 Prokaryotic Initiation Key Features
Initiator tRNA
Initiation also requires the binding of the first tRNA to the ribosome. As we have noted earlier, the initiation or start codon is usually AUG, which codes for the amino acid methionine.
Thus, the initiator tRNA is one that carries methionine and is designated as tRNAmet or methionyl tRNAmet.
In prokaryotes, the methionine on the initiator tRNA is modified by the addition of a formyl group and is designated tRNAfmet.
The initiator tRNA carrying methionine to the AUG is different from the tRNAs that carry methionine intended for other positions in proteins. As such, the initiator tRNA is sometimes referred to as tRNAi-met
fMet is only used for the initiation of protein synthesis and is thus found only at the N-terminus of the protein. Unmodified methionine is used during the rest translation. Once protein synthesis is completed, the formyl group on methionine may be removed and on occasion, the entire methionine residue can be further removed by special enzymes.
Shine-Dalgarno sequence
The ribosome must be correctly positioned at the 5’ end of the messenger RNA in order to initiate translation. How does the ribosome “know” exactly where to bind in the 5’UTR of the mRNA?
Examination of the sequences upstream of the start codon in prokaryotic mRNAs reveals that there is a short purine-rich sequence ahead of the start codon that is crucial to recognition and binding by the small ribosomal subunit.
This sequence called the Shine-Dalgarno sequence, is complementary to a stretch of pyrimidines at the 3’ end of the 16S rRNA component of the small ribosomal subunit
Base-pairing between these complementary sequences positions the small ribosomal subunit at the right spot on the mRNA, with the AUG start codon at the P-site.
Initiation factors
The binding of the small ribosomal subunit to the mRNA requires the assistance of three protein factors called Initiation Factors 1, 2, and 3 (IF1, IF2, IF3).
These proteins, which are associated with the small ribosomal subunit, are necessary for its binding to mRNA but dissociate from it when the 50S ribosomal subunit binds.
IF3= Initiation Factor 3
- An antiassociation factor; prevents association between the large and small ribosomal subunits.
- It also must be associated with the small subunit for it to form an initiation complex, i.e. for the small subunit to correctly bind mRNA and fmet-tRNAf.
- It dissociates prior to binding of the large subunit
IF1 = Initiation Factor 1
- Prevents premature association of amino-acyl tRNAs with the small ribosome subunit
IF2
- Brings fmet‑tRNAf to the partial P site on the small subunit.
- IF2 activates a GTPase activity in the small subunit. The resulting change in conformation may allow the large subunit to bind.
Once the small ribosomal subunit is bound to the mRNA and the initiator tRNA is positioned at the P-site, the large ribosomal subunit is recruited and the initiation complex is formed.
The binding of the 50S ribosomal subunit is accompanied by the dissociation of all three initiation factors.
The removal of IF1 from the A-site on the ribosome frees up the site for the binding of the charged tRNA corresponding to the second codon.
10.5.3 Eukaryotic Initiation- Key Differences from Prokaryotes
The initiation process is slightly more complicated, but the elongation and termination processes are the same, but with eukaryotic homologs of the appropriate elongation and release factors.
Eukaryotic initiation factors are written as – eIFs where the ‘e’ stands for eukaryotic and IF for initiation factor.
Eukaryotes have a large number of IFs involved in the binding of the initiator tRNA to the small subunit, as well as in association of the small subunit with mRNA and subsequent attachment of the large subunit.
We will not cover the action of all the eIFs in detail but rather focus on a few key steps.
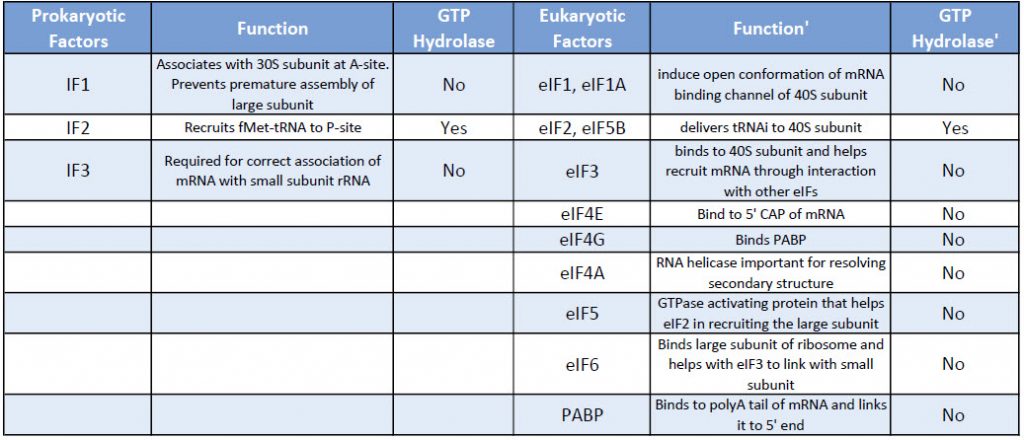
Table: Comparison of Prokaryotic and Eukaryotic Translation Initiation Factors
Key Difference 1: Actively Translating eukaryotic mRNA is circular!
In eukaryotes, the processed mRNA contains additional modifications: A CAP at the 5′ end – bound by CAP binding protein and A Poly-A tail at the 3 end- bound by Poly A Binding Proteins.
This processed mRNA exits the nucleus, and once in the cytoplasm eukaryotic Initiation factors eIF, replace the CAP binding protein one of which is eIF4E.
Another protein eIF4G connects with the eukaryotic initiation factors assembled at the 5′ end with the 3′ end poly-A- binding proteins to create a circular structure. (See Figure 10.11 below)

Figure 10. 11 Eukaryotic Translation Initiation. This is a simplified diagram of eukaryotic translation initiation detailing some of the eIFs involved in the process. eIF2 is critical for recruiting the initiation tRNAi to the 40S subunit. The 43S pre-initiation complex through the interaction of the eIF4 factors and causes the scanning of the pre-initiation complex down the mRNA to locate the start codon (usually AUG). Poly A Binding Proteins (PABPs) bind with the polyA tail sequence of the mRNA and also interact with the eIF4 factors causing the circularization of the mRNA. Figure from: Eukaryotic Translation, Wikiwand
The binding of the mRNA cap by eIF4E is often considered the rate-limiting step of cap-dependent initiation, and the concentration of eIF4E is a regulatory nexus of translational control.
Key Difference 2: Finding the start codon (AUG)
Unlike prokaryotes, the assembly of the translation machinery in eukaryotes begins with the binding of the initiator tRNA to the 40S (small) subunit BEFORE the subunit binds the mRNA.
This step requires the assistance of eIF2 and other factors. The complex of the small ribosome accompanied by eukaryotic initiation factors and Met-tRNAi is known as the ternary complex.
Next, the small complex with the initiator tRNA binds to the 7-methyl G cap on the 5’end of the mRNA.
This 43S preinitiation complex accompanied by the protein factors moves along the mRNA chain toward its 3′-end, in a process known as ‘scanning’, to reach the start codon (typically AUG).
Kozak sequences
Specific sequences surrounding the AUG, called Kozak sequences for the scientist who defined them, have been shown to be necessary for the binding of the 40S subunit, with the bases at -4 and +1 relative to the AUG being especially important.
Once the small subunit is properly positioned, the large ribosomal subunit (60S) binds, forming the initiation complex.
Concepts in Context
Many eukaryotic viruses have enzymes that can remove or clip the eIF4G protein. This prevents eIF4G from binding eIF4E and prevents cap -dependent translation of host messages. Thus these viruses can hijack the host translational machinery in favor of the viral (cap-independent) message!
Watch the lecture video summarizing the information above: Eukaryotic Translation Initiation (17 minutes)
Before you continue
1) Make sure you have watched lecture videos or links to quick review videos provided
10.5.4 Elongation and Termination
Note: The lecture video has additional information on Antibiotics and Translation that is not provided below!
Key Points From Video
We only discuss elongation and termination in a prokaryotic system, due to the similarity between the processes between organisms.
Elongation
After the ribosome is assembled with the initiator tRNA positioned at the AUG in the P-site, the addition of further amino acids can begin.
In both prokaryotes and eukaryotes, the elongation of the polypeptide chain requires the assistance of elongation factors.
In bacteria, the binding of the second charged tRNA at the A-site requires the elongation factor EF-Tu complexed with GTP.
When the charged tRNA has been loaded at the A-site, EF-Tu hydrolyzes the GTP to GDP and dissociates from the ribosome.
The free EF-Tu can then work with another charged tRNA to help position it at the A-site, after exchanging its GDP for a new GTP.
The reaction that joins the amino acids occurs in the ribosomal peptidyl transferase center, which is part of the large ribosomal subunit. This reaction is catalyzed by rRNA components of the large subunit, making the formation of peptide bonds an example of the activity of RNA enzymes, or ribozymes.
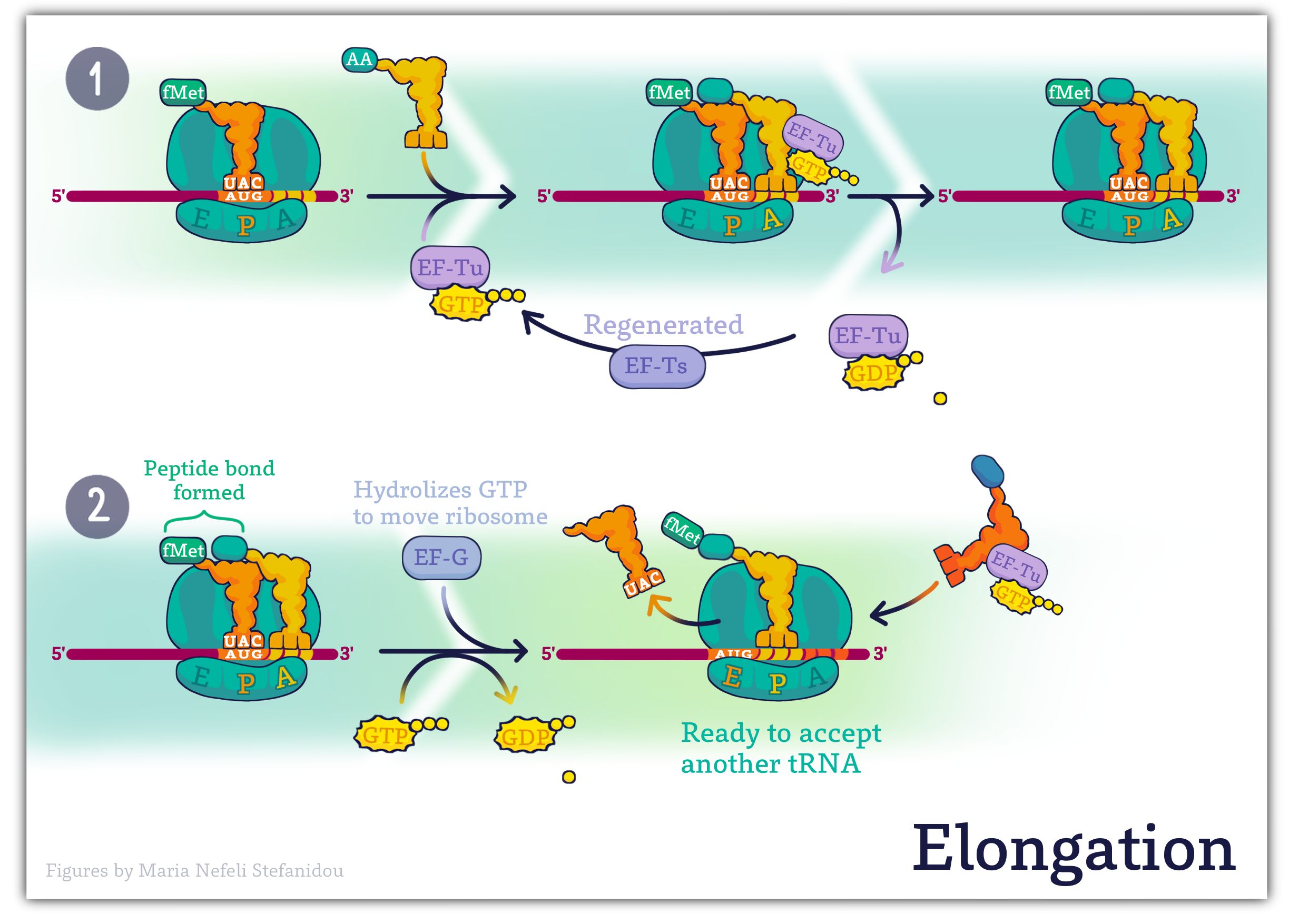
The result of the peptidyl transferase activity is that the tRNA in the A-site now has two amino acids attached to it, while the tRNA at the P-site has none. This “empty” or deacylated tRNA is moved to the E-site on the ribosome, from which it can exit.
The tRNA in the A-site then moves to occupy the vacated P-site, leaving the A-site open for the next incoming charged tRNA.
Another elongation factor, EF-G in complex with the nucleotide GTP, is required for the translocation of the ribosome along the mRNA in bacteria.
Repeated cycles of these steps result in the elongation of the polypeptide by one amino acid per cycle, until a termination, or stop codon is in the A-site.
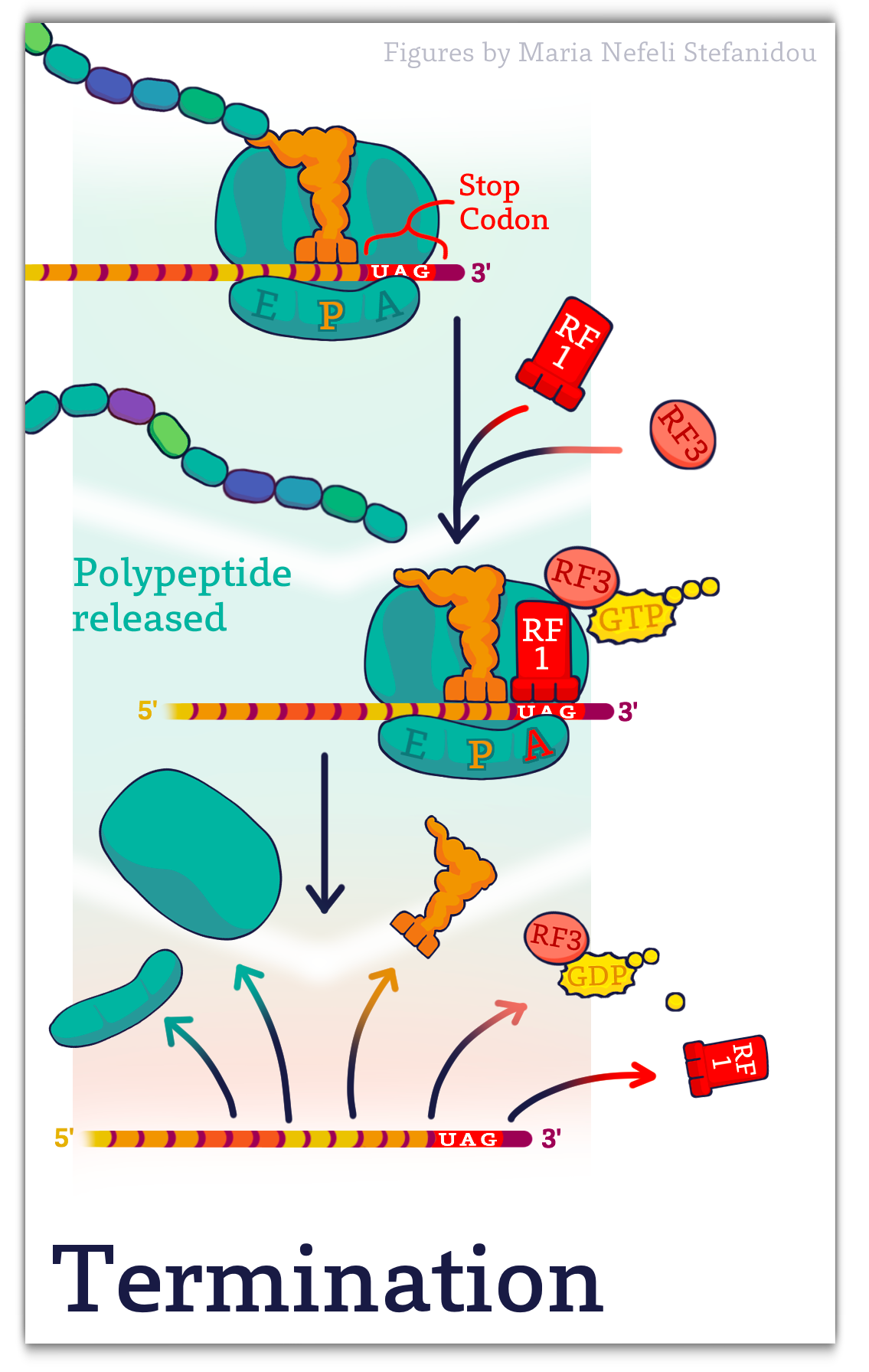
Termination (simplified!)
When a stop codon is in the A-site, Proteins called Release Factors (RF) recognize the stop codon and cleave and release the newly made polypeptide.
In bacteria, RF1 is a release factor that can recognize the stop codon UAG, while RF2 recognizes UGA. Both RF1 and RF2 can recognize UAA. A third release factor, RF3, works with RF1 and RF2 to hydrolyze the linkage between the polypeptide and the final tRNA, to release the newly synthesized protein.
This is followed by the dissociation of the ribosomal subunits from the mRNA, ending the process of translation.
Before you continue you should
1. Watch any videos linked above
Answers to Problem in text:
[modifications of mRNA ; (b) ]
References and Attributions
This chapter contains material taken from the following CC-licensed content. Changes include rewording, removing paragraphs and replacing with original material, and combining material from the sources.
1. Bergtrom, Gerald, “Cell and Molecular Biology 4e: What We Know and How We Found Out” (2020). Cell and Molecular Biology 4e: What We Know and How We Found Out – All Versions. 13.
https://dc.uwm.edu/biosci_facbooks_bergtrom/13
2. Works contributed to LibreTexts by Kevin Ahern and Indira Rajagopal. LibreTexts content is licensed by CC BY-NC-SA 3.0. The entire textbook is available for free from the authors at http://biochem.science.oregonstate.edu/content/biochemistry-free-and-easy
3. Flatt, P.M. (2019) Biochemistry – Defining Life at the Molecular Level. Published by Western Oregon University, Monmouth, OR (CC BY-NC-SA). Available at: https://wou.edu/chemistry/courses/online-chemistry-textbooks/ch450-and-ch451-biochemistry-defining-life-at-the-molecular-level/?preview_id=4919&preview_nonce=cca8f0ce36&preview=true
4. “Translation” by Katherine Harris, LibreTexts is licensed under CC BY-NC-SA .
5. “Protein Synthesis (Translation)” by OpenStax, LibreTexts is licensed under CC BY .