Molecular Biology: From DNA to RNA to Protein
9 mRNA Splicing and Alternative Splicing
Learning Objectives
- Know the similarities and differences that exist between pre-mRNA and mRNA.
- Know/draw/label the steps involved in spliceosome formation.
- Describe the roles of snRNPs in splicing
- Explain what are the benefits of RNA processing.
- What is transesterification, and what transesterification reactions are needed to splice introns?
- Explain why fidelity of splicing is important.
- Explain what controls the fidelity of splicing.
- Predict how mutations at the 5’ splice site, 3’ splice site, and branch point might disrupt splicing and alter the phenotype.
- Describe the different forms/patterns of alternate splicing.
- What does constitutive exon mean?
- Draw splicing diagrams to show alternative splicing patterns.
- Identify when given a description/diagram of an alternately spliced gene which type it is.
- How is alternative splicing regulated?
- What are exonic and intronic splicing silencers or enhancer sequences? (ESS, ISS, ESE, and ISE)
- Using examples explain how anti-sense technology is used to correct for splicing defects.
CONNECTING CONCEPTS
Chapters 8 and 9 introduced you to different components of eukaryotic gene structure and RNA molecules transcribed. These include promoters, 5′ and 3′ untranslated regions (UTR), coding sequences (exons), introns, 5′ caps, Poly A signal, and poly(A) tails.
You should be able to draw /identify/annotate when given a gene sequence the elements of the gene above.
You should be able to draw the pre-mRNA and mRNA derived from it.
9.1 Introduction
The immediate product of RNA polymerase II is sometimes referred to as pre-mRNA or the primary transcript.
As we saw in Chapter 8, the initial products of transcription are further processed acquiring a cap at their 5′ end and poly-A tail at their 3′ end. Most importantly nearly ALL mRNA precursors are spliced
Splicing is the process by which the non-coding regions, known as introns, are removed, and the coding regions, known as exons, are connected together.
Introns were initially thought to be entirely a feature of the eukaryotic genome. In recent years the presence of intron-containing genes has been documented in archaea, bacteriophages, and even some bacteria. They are also present within mitochondrial and chloroplast genes. These introns are called Group I and II introns and are self-splicing!
That means for those RNAs, splicing happens autonomously, with part of the RNA acting as an enzymatic catalyst for the process. This requires that the RNA have a specific secondary and tertiary structure, bringing the two exons close together while looping out the intron. It was the study of this phenomenon that led to the discovery of ribozymes, which are enzymes made of RNA.
Until the discovery of ribozymes, it had been assumed that enzymes could only be generated with the diversity of structures possible with the amino acids in proteins.
The splicing process we will study however is carried out within the nucleus on mRNA using a multi-subunit protein complex known as the Spliceosome.
Spliceosome-mediated splicing IS a unique feature of eukaryotes.
9.2 The Split Gene
Discovery of Interrupted Genes
The discovery of eukaryotic split genes with introns and exons came as quite a surprise.
Go to the DNA Learning Center website and click on the Interactive Animation that outlines the experiments that led to the discovery that eukaryotic genes have non-coding regions.
For their discovery of split genes, Richard J. Roberts and Phillip A. Sharp shared the Nobel Prize for Physiology in 1993.
In fact, all but a few eukaryotic genes are split, and some have one, two (or more than 30-50!) introns separating bits of coding DNA, the exons. The size of the intron (# of base pairs) and number of introns may vary.
An extreme example of splicing and of medical relevance is the human dystrophin gene.
Human Dystrophin Gene- Muscular Dystrophy
The DMD gene is one of the largest known gene in humans, spanning 2.6 million base pairs (bp) consisting of almost 0.1% of the human genome or about 1.5% of the entire X chromosome. This very large gene is highly fragmented into 79 exons and introns of variable size ranging from 107 bp (intron 14) to >200kb (intron 44).
It is transcribed in a 14 kb mRNA, and the 11kb cDNA encodes a 3685 amino acid protein of 427 kDa called Dystrophin.
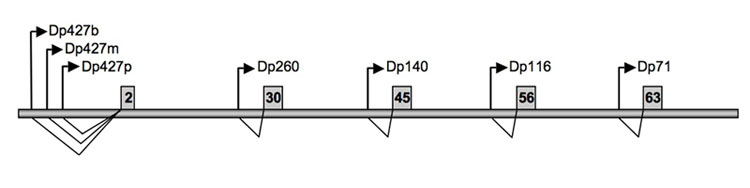
Dystrophin is located primarily in muscles used for movement (skeletal muscles) and in heart (cardiac) muscle. Small amounts of dystrophin are present in nerve cells in the brain.
Many different types of mutations have been described for DMD including large deletions and duplications, point mutations, and small rearrangements that underlie various forms of Muscular Dystrophy.
9.2.1 So, Why Splicing?
While the dystrophin gene is an extreme example, overall in humans
a) Median Gene is about 23, 000 bp, 7 introns
b) Median Intron is about 1800 bp in size and
c) Median Exon is only about 123 bp in size!
We can see that 90 -95% of the RNA transcript that gets synthesized is essentially thrown out! So why do higher organisms have split genes in the first place?
While the following discussion can apply to all splicing, it will reference mainly spliceosomal introns. Here are some answers to the question “Why splicing?
1 ) Introns in nuclear genes are typically longer (often much longer!) than exons. Since they are non-coding, they are large targets for mutation. In effect, noncoding DNA, including introns can buffer the ill effects of random mutations.
2) Gene duplication on one chromosome (and loss of a copy from its homolog) arises from unequal recombination (non-homologous crossing over). Unequal recombination can also occur between similar sequences (e.g., in introns) in the same or different genes, resulting in a sharing of exons between genes.
After unequal recombination between introns flanking an exon, one gene will acquire another exon while the other will lose it. Oftentimes times the new gene will produce the same protein, but one with a new structural domain and function. Thus, this phenomenon of exon shuffling increases species diversity! If the protein with a new domain is beneficial then natural selection might increase the prevalence of this protein.
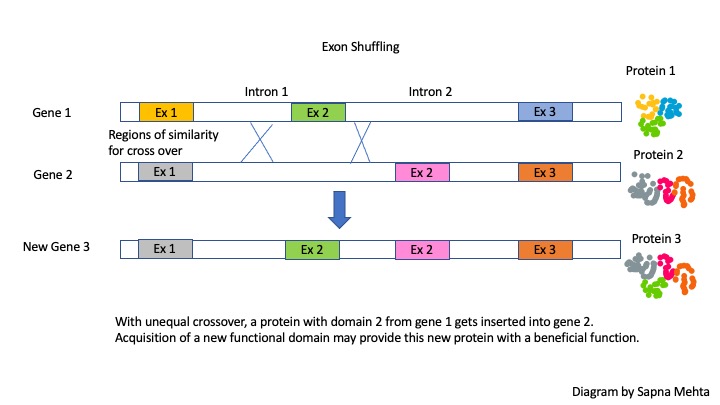
Many existing proteins are predicted to have arisen via exon shuffling, creating proteins with different overall functions that nonetheless share at least one domain and one common function.
3) The presence of introns allows for increasing protein diversity. As shown in Figure 9.1 one gene with 5 exons can produce at least 3 different isoforms as illustrated, depending on which exons are joined together.
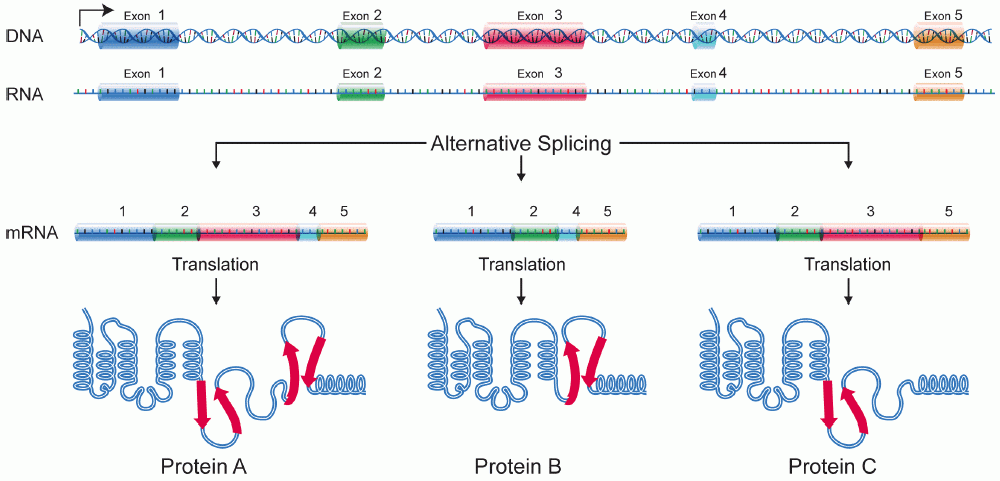
9.3 A Detailed Look at mRNA Splicing
Splicing must be exquisitely sensitive because the nucleotide information is converted into protein information in three non-overlapping nucleotide sequences called codons (See Themes from Inro Bio and Chapter on Translation). Thus, a one-nucleotide shift in the course of splicing would alter the nucleotide information on the processed mRNA resulting in inaccurate coding sequence and protein.
Therefore the splicing machinery must be able to recognize splice junctions (i.e., where each exon ends and its associated intron begins) in order to correctly cut out the introns and join the exons to make the mature, spliced mRNA.
9.3.1 Finding the Intron/Exon Junctions
Whether it is self-spliced or using the spliceosome, the junctions between exons and introns are indicated by specific base sequences and guide the splicing process called SPLICE SITES.
They are named for their positions relative to the intron. These include
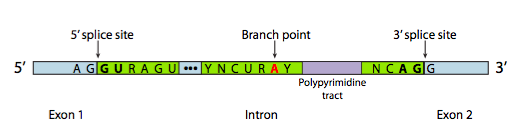
- A 5’ splice site with the consensus sequence AG|GURAGU.
In this sequence, the intron starts
with the second G (R stands for any purine)
2. A 3’ splice site that starts with an 11-nucleotide polypyrimidine tract followed by NCAG|G.
…and somewhere in between the two,
3. A branchpoint adenine, typically within a YNCURAY sequence (Y is a pyrimidine, N is any nucleotide, R is a purine)
The branch point Adenine is ‘invariant’- meaning it is always needed and present in introns. The importance of this site will be seen when we consider the steps of splicing.
Notice that the INTRON is defined by a GU at its 5′ end and an AG at its 3′ end. Splicing occurs utilizing sets of GU-AG marking the boundary of the intron at either end where the ‘cut/ splice’ needs to occur.
This is often referred to as the GU-AG rule: (originally called the GT-AG rule in terms of DNA sequence) describing the requirement for these constant dinucleotides at the first two and last two positions of introns in pre-mRNAs.
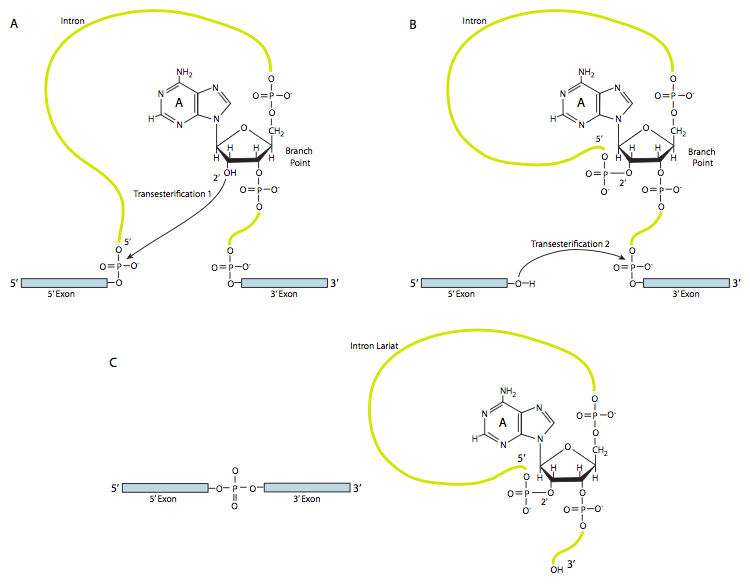
9.3.2 The Chemistry of Splicing: Transesterification
There are two main steps in splicing.
STEP 1: The nucleophilic attack by the 2’OH of the branch point A on the 5′ splice site (the junction of the 5′ exon and the intron), releasing the 5′ exon with a free 3′ hydroxyl group. A looped lariat-shaped molecule composed of the 5’ end of the intron connected to the branchpoint via a 2’,5’-phosphodiester bond.
We can now see why BRANCH point A is crucial for splicing to occur.
As a result of a trans-esterification reaction, the 5′ exon is released, and a lariat-shaped molecule composed of the 3’ exon and the intron sequence is generated (Figure 9.3). The lariat 5’ end of the intron is connected to the branchpoint via a 2’,5’-phosphodiester bond.
STEP 2: The 3′ OH of the 5’ exon which was released in Step 1, attacks the phosphate at the 3’ splice junction, connecting exons 1 and 2. The lariat intermediate is released.
9.3.3 The Spliceosome
More than 300 proteins and a group of special RNAs come assembled on the pre-mRNA to form the machine called the ‘Spliceosome’ that controls mRNA splicing.
The components of this machine include small nuclear ribonucleoproteins or snRNPs (pronounced “snurps”) for short.
As the name suggests, snRNPs contain proteins and a small nuclear RNA (snRNA) component. The snRNAs are designated as U1, U2, U4, U5, and U6. These are 100-200 nucleotides long and adopt elaborate tertiary structures.
They associate with many proteins and together form the snRNP. The snRNPs are named after the RNA component, for example, the U1 snRNP consists of many proteins along with a U1 snRNA component.
There are therefore the U1 snRNP, U2 snRNP, U4 snRNP, U5 snRNP, and U6 snRNP. The snRNPs recognize the conserved sequences within introns and quickly bind these sequences once the pre-mRNA is made and initiate splicing.
It is the RNA component that base pairs with the consensus site and brings the snRNP to the accurate location.
Although many details remain to be worked out, it appears that components of the splicing machinery associate with the CTD of the RNA polymerase and that this association is important for efficient splicing.
Other proteins that play a role include U2AF (U2-associated factor, which binds to the polypyrimidine tract, and branch-point protein BPP, which binds to the consensus sequence near the branchpoint.
There are also a variety of other less-studied splicing factors from the SR protein family (C-terminal Serine-Arginine binding motif) and the hnRNP (heterogeneous nuclear ribonucleoprotein) families that act to recruit the primary members of the spliceosome to their proper locations (see Alternative splicing regulation)
The spliceosome is not ‘pre-assembled’ but builds on the mRNA in a step-wise fashion as the mRNA emerges from the RNA polymerase.
Key Takeaways
Practice Questions
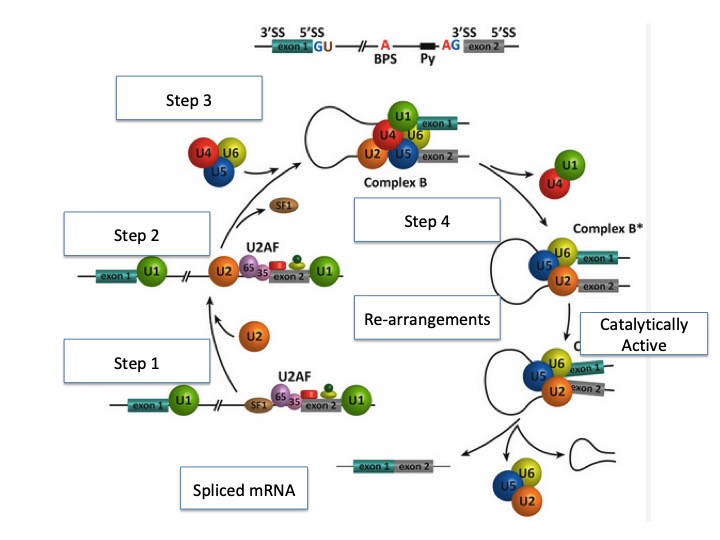
9.3.4 Steps in splicing
We will illustrate the assembly of the spliceosome using an example transcript. In this example, the pre-mRNA contains two exons and one intron. (Figure 9. 4)
First, the U1 snRNP binds the 5’ splice site. Additional proteins such as U2AF (AF = associated factor) are also loaded onto the
pre-mRNA near the branch site.
Next, the U2 snRNP binds to the consensus site around the branchpoint, but importantly, there is no base pairing to branchpoint A itself. Instead, due to the base pairing of U2 with the surrounding sequence, the branch point A is forced to bulge out from the rest of the RNA in that region.
Next, a complex of the U4/U6 and U5 snRNPs is recruited to the spliceosome to generate a pre-catalytic complex. This com-
plex undergoes rearrangements that alter RNA-RNA and protein-RNA interactions, resulting in the displacement of the U4 and U1 snRNPs and the formation of the catalytically active spliceosome.
This complex is responsible for executing the two transesterification steps: First, the 5’ end of the intron is cut. The 5’ GU end of the intron is then connected to the A branch site, which creates a lariat structure. This releases the 5’ exon (and the whole 5’ half of the RNA, for that matter), but it is kept near the 3’ exon (and the rest of the RNA) by U5, which attaches to both exons.
This allows the second transesterification to take place, in which the 3’- OH of the first exon attacks the 5’ phosphate at the beginning of the second exon, simultaneously breaking the bond between the intron and the second exon and connecting the two exons via a conventional 3’,5’-phosphodiester bond.
After the second catalytic step, the intron is released as a lariat along with U2, U5, and U6 snRNPs.
The intron will be degraded, and the snRNPs will be used again to splice other pre-mRNAs. The mature mRNA transcript is now ready to be exported to the cytoplasm for translation.
Once splicing is complete, proteins called Exon Junction Complexes (EJC) are deposited, marking the previous boundaries.
Along with the CAP-binding protein at the 5′ end and the poly at tail binding protein at the 3′ end, the presence of these protein markers indicates a processed and mature mRNA transcript that is ready for export out of the nucleus into the cytoplasm, where it will be translated into protein.
9.4 Alternative Splicing
Splicing is an efficient (for genome size) way to generate protein diversity. In alternative splicing, some potential introns may be spliced out under certain circumstances but remain as coding sequences under other circumstances.
Alternative Splicing (AS) thus offers an additional mechanism for regulating protein production and function.
Recall that the splice sites are recognized by base-pairing, and therefore, there can be stronger and weaker splice sites depending on how close they are to the consensus and the complementary sequence of the snRNPs. Therefore, a gene with several potential introns may have all introns spliced out 80% of the time, but the other 20% of the time, perhaps only one or two introns are spliced out.
Adding variability, splicing factors may bind near splice sites and can either make them more easily recognizable or nearly hidden (see Alternative Splicing Regulation below).
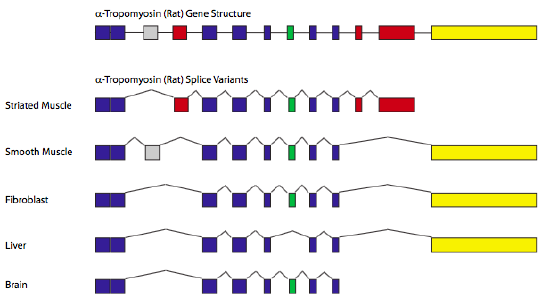
A classic example of alternative splicing is the gene encoding α-tropomyosin.
By splicing in/out different combinations of exons, a single gene can generate seven different proteins, depending on the tissue type (Figure 9.5)
9.4.1 Alternative Splicing Terminology
Splice forms /Splice variants/ Isoforms: Refers to all the different ways in which a pre-mRNA can be spliced to generate different forms of mature mRNA
CONSTITUTIVE EXON: Those exons that are always included in all splice forms
REGULATED EXONS: Those that are sometimes included, sometimes not.
9.4.2 Alternative Splicing Patterns
I. Exon skipping: This is the major alternative splicing event in higher eukaryotes. In this type of event, the exon and the intron flanking on either side are removed from the pre-mRNA (Fig. 9.6 a).
II. Alternative 3′ and 5′ SS selection (Fig. 9.6 b & c) or Exon Extension: Occurs when the spliceosome recognizes two or more (an alternative) splice sites at one end of an exon.
In the diagram below, starting from the left and moving into the mRNA, the spliceosome uses the GU-AG rule. In the 3′ SS selection, the spliceosome pairs the 5′ SS with an alternative 3’SS. Conversely, an alternative 5′ SS pairs with a normal 3’SS.
Using alternative 3’SS and 5′ SS also results in exon extension.
III. Intron retention (Fig. 9.6 d): in which an intron remains in the mature mRNA transcript. This alternative splicing event is much more common in plants, fungi, and protozoa than in vertebrates.
Other events that affect the transcript type (isoform) outcome include mutually exclusive exons (Fig. 9.6 e), alternative promoter usage (Fig. 9.6 f), and alternative polyadenylation (Fig. 9.6 g).
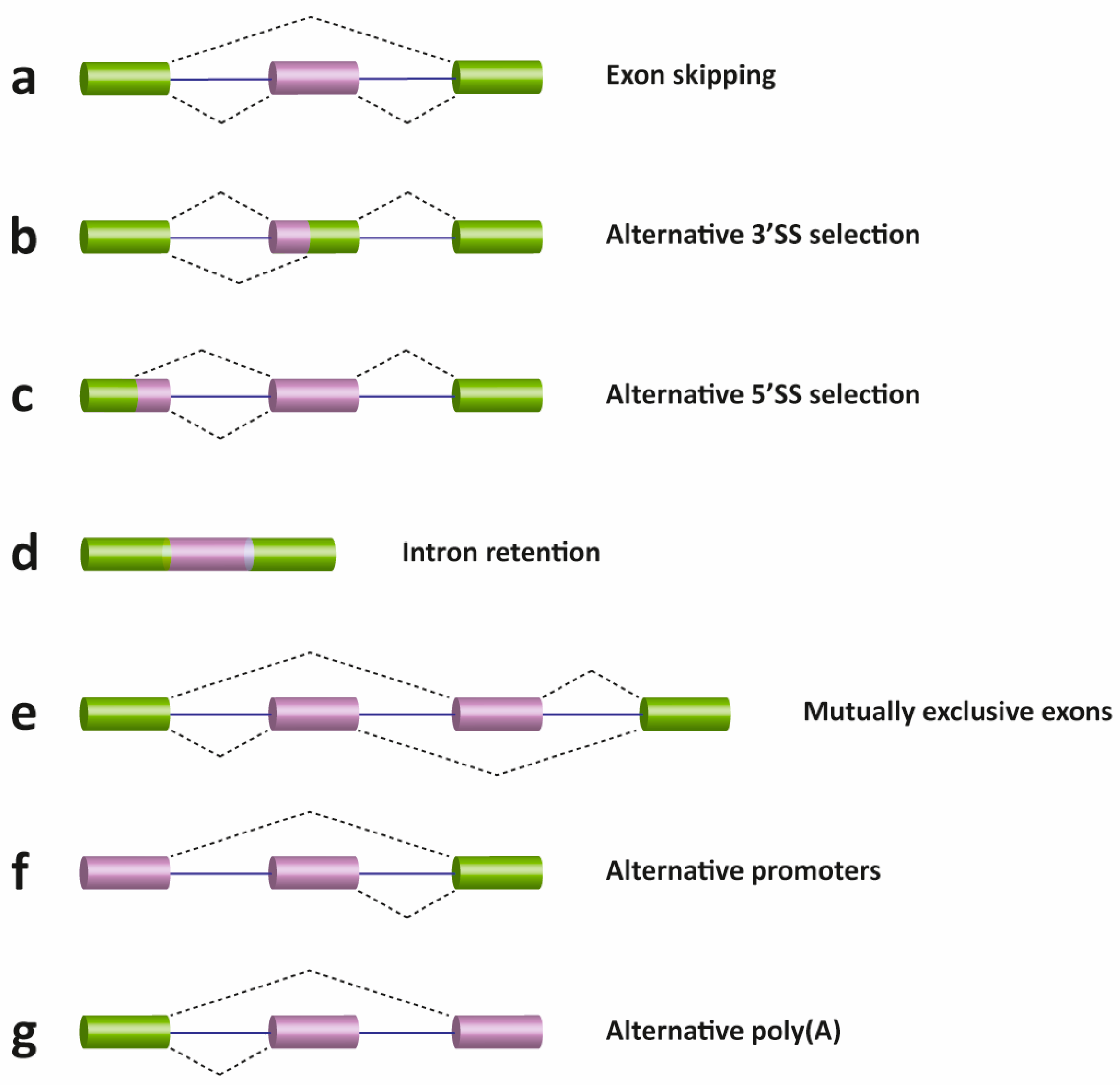
Figure 9.6 Schematic representation of different types of alternative transcriptional or splicing events, with exons (boxes) and introns (lines). Constitutive exons are shown in green and alternatively spliced exons in purple. Dashed lines indicate the AS event. Exon skipping (a); alternative 3′ (b) and 5′ SS selection (c); intron retention (d); mutually exclusive exons (e); alternative promoter usage (f); and alternative polyadenylation (g) events are shown. Like alternative splicing (AS), usage of alternative promoter and polyadenylation sites allow a single gene to encode multiple mRNA transcripts.
Figure from: Suñé-Pou, M., et. al. (2017) Genes 8(3):87
Examples: Alternative Polyadenylation
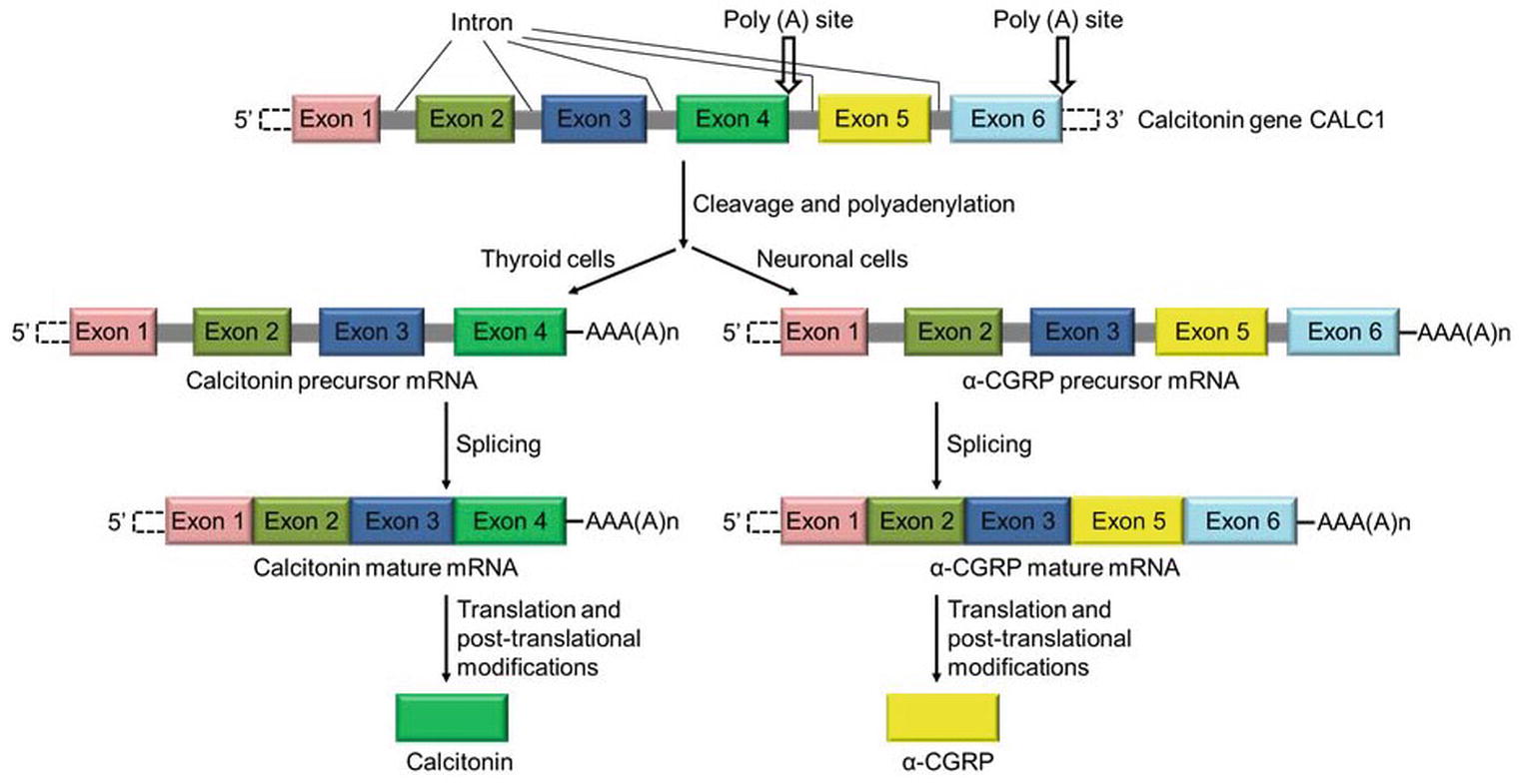
The CT/CGRP gene is an example of alternative splicing using an alternative poly(A) site. The gene produces the same pre-mRNA transcript in many cells, including thyroid cells and neuronal cells. The transcript contains six exons and five introns and includes two alternative polyadenylation sites, one in exon 4 and the other following exon 6.
In thyroid C cells, the gene product contains exon 4. The inclusion of Exon 4 facilitates utilization of the Poly(A) site and cleavage of mRNA transcript and termination of transcription. Translation of this mRNA forms calcitonin, a hormone that regulates calcium in the body.
In sensory neuronal cells, the same pre-mRNA is spliced to include exons 1,2,3 5, and 6, skipping exon 4. Therefore Poly-adenylation takes place at the site in Exon 6. The mature mRNA when translated forms α-CGRP a regulatory neuropeptide.
9.4.3 Alternative Splicing Regulation
Alternative splicing is not a random process and is controlled by regulatory proteins. These proteins bind to the pre-mRNA and “tell” or “influence” the splicing factors which splice sites to engage in.
These regions on the pre-mRNA that proteins bind are known as exonic and intronic splicing silencers or enhancers (ESS, ISS, ESE, and ISE, respectively).
(Not to be confused with enhancer sequences found on DNA that influence transcription!)
Specific RNA-binding proteins, including heterogeneous nuclear ribonucleoproteins (hnRNPs) and serine/arginine-rich (SR) proteins, recognize these sequences to positively or negatively regulate alternative splicing (Figure 9.7).
In general, SR proteins bound to enhancers facilitate exon definition, and hnRNPs inhibit this process. These trans-acting elements are expressed differentially within different locations or under different environmental stimuli to regulate alternative splicing.
These regulators and an ever-increasing number of additional auxiliary factors provide the basis for the specificity of this pre-mRNA processing event in different cellular locations within the body.
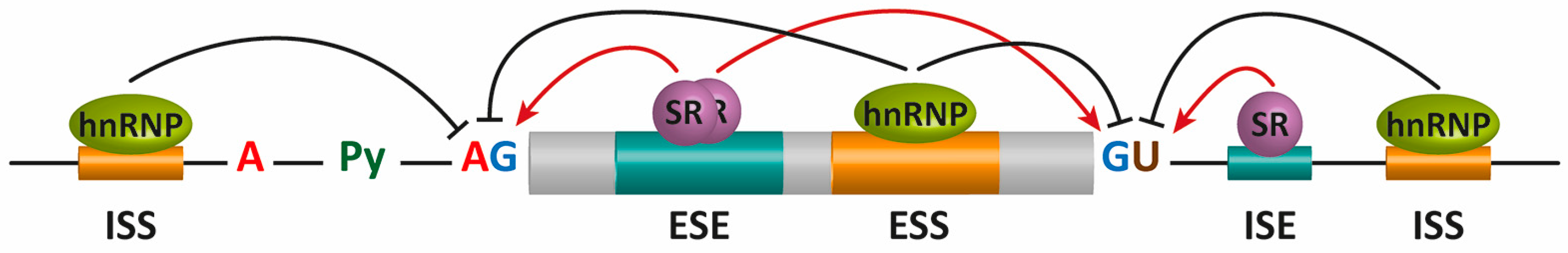
Figure 9.7 Alternative splicing (AS) regulation by cis mRNA elements and trans-acting factors. The core cis sequence elements that define the exon/intron boundaries (5′ and 3′ splice sites (SS), GU-AG, polypyrimidine (Py) tract, and branch point sequence (BPS)) are poorly conserved. Additional enhancer and silencer elements in exons and in introns (ESE: exonic splicing enhancers; ESI: exonic splicing silencers; ISE: intronic splicing enhancers; ISI: intronic splicing silencers) contribute to the specificity of AS regulation. Figure from: Suñé-Pou, M., et. al. (2017) Genes 8(3):87
Check your understanding
9.5 Clinical Insight
Many human genetic diseases arise from mutations that affect pre-mRNA splicing; indeed, about 15% of single-base substitutions that result in human genetic diseases alter pre-mRNA splicing. Some of these mutations interfere with the recognition of the normal 5′ and 3′ splice sites. Others arise from mutations in splicing factors that can also cause defective mRNA splicing.
Listed below are some common diseases and the associated splicing defects.
| Disease | Mutation | Splicing Effect |
|---|---|---|
| Familial dysautonomia (FD) | T > C mutation at position 6 of intron 20 of the IKBKAP gene | Exon skipping; introduction of a premature termination codon (PTC) |
| Spinal muscular atrophy (SMA) | C > T mutation at position 6 of exon 7 of the SMN2 gene | Alteration of a putative ESE |
| Hutchinson-Gilford progeria syndrome (HGPS) | c1824C > T mutation in exon 11 of LMNA gene | Activation of a cryptic splice site |
| Duchenne muscular dystrophy (DMD) | T > A mutation in exon 31 of the Dystrophin gene | Creation of a PTC and introduction of ESS |
| Amyotrophic lateral sclerosis (ALS) | Mutations in TDP-43 | Altered gene splicing |
| Autosomal dominant retinitis pigmentosa (RP) | Mutations in genes of the core spliceosome (PRPF31, PRPF8, PRPF3, RP9) | Disruption of basal spliceosome function |
9.5.1 Antisense Oligonucleotide Therapy
Concepts in Context: Molecular Biology in the News
In December 2016 the FDA approved Spinraza, the first FDA-approved therapy for all ages and types of SMA- Spinal Muscular Atrophy. SMA affects approximately 1 in 11,000 births in the U.S., and about 1 in every 50 Americans is a genetic carrier. SMA can affect any race or gender.
This discovery came after years of basic research to understand splicing and the mechanisms behind it. It was conceived and tested over several years in mouse models of SMA by Professor Adrian Krainer, Ph.D., and his colleagues at Cold Spring Harbor Laboratory (CSHL)
Watch this video on the mechanism of action of Spinraza
Spinraza is one of a growing list of gene therapies using an antisense oligonucleotide (ASO) for modifying and fixing errors in the splicing process. Antisense drugs are small snippets of synthetic genetic material that bind to ribonucleic acid (RNA), so they can be used to fix the splicing of genes like SMN2.
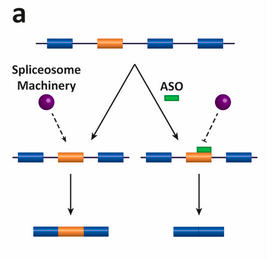
The diagram depicts the antisense oligonucleotide (ASO)–based strategy for altering splicing patterns. Here the antisense oligos target an alternatively spliced exon (in orange). In the absence of the ASO, the spliceosome is assembled and the exon is included in the mRNA; in the presence of the ASO, the spliceosome is sterically blocked and the exon is skipped and not included in the mRNA.
Before you continue
- Complete the Lecture Quick checks associated with this unit.
- Complete the associated Concepts in Context Assignment.
References and Attributions
This chapter contains material taken from the following CC-licensed content. Changes include rewording, removing paragraphs and replacing them with original material, and combining material from the sources.
1. Section Heading and content “So, Why Splicing” from Bergtrom, Gerald, “Cell and Molecular Biology 4e: What We Know and How We Found Out” (2020). Cell and Molecular Biology 4e: What We Know and How We Found Out – All Versions. 13.
https://dc.uwm.edu/biosci_facbooks_bergtrom/13
2. Works contributed to LibreTexts by Kevin Ahern and Indira Rajagopal. LibreTexts content is licensed by CC BY-NC-SA 3.0. The entire textbook is available for free from the authors at http://biochem.science.oregonstate.edu/content/biochemistry-free-and-easy
3. Flatt, P.M. (2019) Biochemistry – Defining Life at the Molecular Level. Published by Western Oregon University, Monmouth, OR (CC BY-NC-SA). Available at: https://wou.edu/chemistry/courses/online-chemistry-textbooks/ch450-and-ch451-biochemistry-defining-life-at-the-molecular-level/?preview_id=4919&preview_nonce=cca8f0ce36&preview=true
Other References
Suñé-Pou, M., Prieto-Sánchez, Boyero-Corral, S., Moreno-Castro, C., El Yousfi, Y., Suñé-Negre, J.M., Hernández-Munain, C., and Suñé, C. (2017) Targeting splicing in the treatment of human disease. Genes 8(3):87. Available at: https://www.mdpi.com/2073-4425/8/3/87/htm

