Molecular Biology: From DNA to RNA to Protein
5 DNA Replication
5.1 Introduction
A fundamental property of living organisms is their ability to reproduce. Bacteria and fungi can divide to produce daughter cells that are identical to the parental cells. Sexually reproducing organisms produce offspring that are similar to themselves.
On a cellular level, this reproduction occurs by mitosis, the process by which a single parental cell divides to produce two identical daughter cells.
Meiosis is the process in which cells with a diploid genome produce four germ cells (haploid cells). Regardless of the type of cell division, the duplication of all of the genetic material is needed so that each daughter cell receives a full complement of genetic material.
In this chapter, we look at the details of replication as well as differences in detail between prokaryotic and eukaryotic replication that arise because of differences in DNA packing.
We will begin with a discussion on the general features of replication common to the replication of ‘naked’ prokaryotic DNA and of chromatin-encased eukaryotic DNA, arisen early in the evolution of replication biochemistry.
You will read about experiments that explored or revealed answers to some basic questions like:
- Where does replication begin? Is it one region on the chromosome or several?
- Is the start random or at a specific location?
- Once replication begins in which direction does replication occur?
Answers to many of these questions arose from experiments carried out in E. coli, which has a circular genome.
We will then introduce all the proteins involved in carrying out this essential process to develop a complete picture of how DNA replication actually occurs, ending with differences between prokaryotic and eukaryotic replication that arise because of differences in DNA packing.
Learning Objectives
Levels 1 and 2 are factual information, and knowledge-based. Level up indicated by the “Target” symbol is the goal.
When you have mastered the information in this chapter, you should be able to:
Levels 1 and 2 (Knowledge and Comprehension)
- Be able to explain why DNA replication is semiconservative
- Be able to explain the data obtained by Meselson-Stahl to prove that replication is semi-conservative.
- Outline the general feature of replication that are common amongst prokaryotes and eukaryotes
- Accurately label within a replication fork (a) the polarity of the newly synthesized strands (5’-3’), (b) leading and lagging strands.
- Identify individual proteins and their roles in DNA replication.
- Why do eukaryotes need telomeres, but prokaryotes do not?
- What are the ways in which bacterial and eukaryotic replication differ?
- What are the ways in which bacterial and eukaryotic replication are similar?
- Explain how eukaryotic cells distribute their histones during replication and what impact it has on chromatin state inheritance.
⊕ Level Up (Application, Analysis, Synthesis)
- What are the requirements for in vitro synthesis of DNA under the direction of DNA polymerase I?
- Apply the concepts of directionality of DNA synthesis to new modes of replication.
- Draw/Interpret modes of replication when given fictitious Meselsohn and Stahl data.
- Predict the effect on DNA replication with mutations in components of DNA replication machinery.
- Predict which component of DNA replication machinery is mutated, when given descriptions of phenotypes of mutants.
5.2 The Basic Rules of Replication
DNA Replication is essential for life and as a process, it must be
- Accurate (make few mistakes)
- Fast
- Complete (although we will see one exception when it comes to the ends of linear chromosomes.)
It takes an army of proteins and some specialized DNA sequences that function together to accomplish this!
While the individual components are important, it is important to not lose sight of some general principles of replication that are common to all of life.
5.2.1 DNA Replication is always semiconservative: Meselson and Stahl Experiments
Watson and Crick’s double helix model suggested a replication mechanism. In presenting the complementary pairing of bases in the double helix, Watson and Crick immediately realized that the base sequence of one strand of DNA can be used as a template to make a new complementary strand.
This was the semi-conservative model of replication. However, how DNA can separate (denature) was not understood and there were alternative models of replication (Figure 5.1)
Conservative Model: Perhaps there is some DNA synthesizing machine in the cell that can take dsDNA and make a copy of it ?
Dispersive Model: Perhaps the process of replication could break the parental DNA into pieces and use them to seed the synthesis of new DNA?
It was far from obvious what the mechanism would be. However, these three models make different predictions about the behavior of the two
strands of the parental DNA during replication.
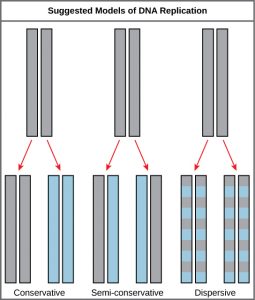
Meselson and Stahl’s set out to test the three possible models of replication and confirmed that DNA replication is in fact semi-conservative. (Due to the simplicity and indisputable results of the experiment, the Meselson-Stahl experiment has been called “the most beautiful experiment in biology.” see Link to Learning)
In their experiment, E. coli cells were grown in a medium containing 15N, a ‘heavy’ nitrogen isotope. After many generations, all of the DNA in the cells had become labeled with the heavy isotope. At that point, the 15N-tagged cells were placed back in a medium containing the more common, ‘light’ 14N isotope and allowed to grow for exactly one generation.
Fig. 5.2 (below) shows Meselson and Stahl’s predictions for their experiment. Meselson and Stahl knew that 14N-labeled and 15N-labeled DNA would form separate bands after centrifugation on CsCl chloride density gradients.
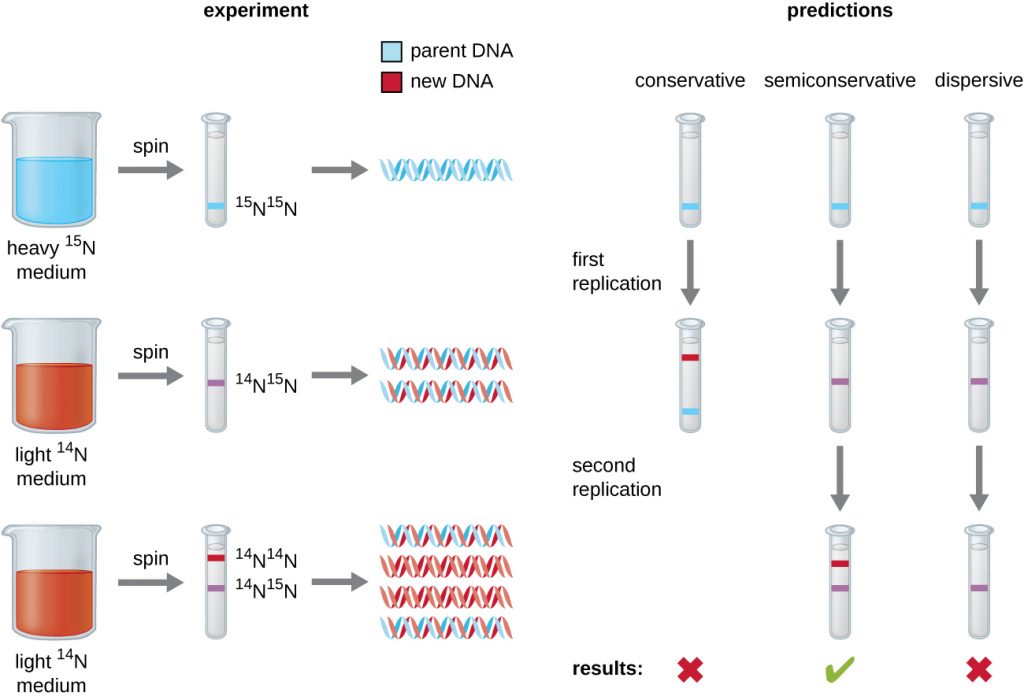
They tested their predictions by purifying and centrifuging the DNA from the 15N-labeled cells grown in 14N medium for one generation.
They found that this DNA formed a single band with a density between that of 15N-labeled DNA and 14N-labeled DNA, eliminating the conservative model of DNA replication (possibility #1).
That left two possibilities: replication was either semiconservative (possibility #2) or dispersive (possibility #3).
The dispersive model was eliminated when DNA isolated from cells grown for a 2nd generation on 14N were shown to contain two bands of DNA on the CsCl density gradients.
iBiology Video: The most beautiful experiment in science
Talk Overview
Matt Meselson and Frank Stahl were in their mid-20s when they performed what is now recognized as one of the most beautiful experiments in modern biology. In this short film, Matt and Frank share how they devised the groundbreaking experiment that proved semiconservative DNA replication, what it was like to see the results for the first time, and how it felt to be at the forefront of molecular biology research in the 1950s. This film celebrates a lifelong friendship, a shared love of science, and the serendipity that can lead to foundational discoveries about the living world.
Exercises
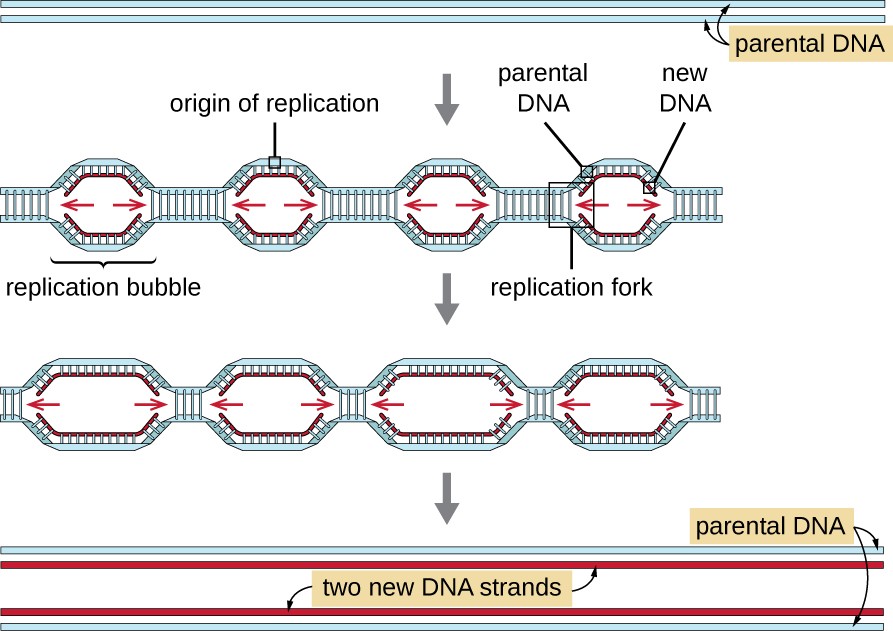
5.2.2 DNA Replication Begins at Specific Chromosomal Sites
Where does replication begin? Does the DNA unwind at one region on the genome or in multiple places? Is it random or are there special sequences on the DNA where the process of unwinding begins?
The answers to some of the fundamental questions came from the direct visualization of bacterial DNA using a technique called autoradiography– an imaging technique utilizing radioactive sources contained within the exposed sample.
Cairns Experiment
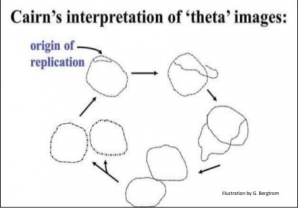
In 1963, John Cairns cultured E. coli cells for long periods on 3H-thymidine (3H-T) to make all of their cellular DNA radioactive. He then extracted the DNA (during replication) and allowed it to adhere to membranes.
A sensitive film was placed over the membrane and time was allowed for the radiation to expose the X-ray film. As the radioactive substance decays the emissions create a pattern on the x-ray film. When the film is developed an image of the pattern is seen, similar to the light that exposes a photographic film. The image obtained is an autoradiograph.
The image pictured in Figure 5.3 shows dark lines (tracks of silver grains in the autoradiographs) that revealed the pattern of replicating DNA molecules.
Cairns called these replicating chromosomes theta images because they resembled the Greek letter theta (θ).
From his many autoradiographs, he arranged a sequence of his images to illustrate his inference that replication starts at a single origin of replication on the bacterial chromosome, proceeding around the circle to completion.
5.2.3 Most DNA Replication Is Bidirectional
The theta images obtained above do not distinguish between uni-directional or bi-directional replication forms of replication for the bacterial chromosome.
In Uni-directional replication- a new DNA molecule is synthesized in one direction only. DNA unwinds at ONE replication fork and moves around the entire circular DNA until complete.
In Bi-directional replication- there are 2 growing points, DNA unwinds, and new DNA is synthesized at BOTH replication forks until they meet at the opposite end from where replication began (the origin).
Additional experiments demonstrated that DNA replication is bi-directional.
- Cells were labeled with 3H-thymidine with low specific activity to lightly label the replication bubble.
- Then the cells were labeled with a much stronger radioactive isotope for a short time.
- Any newly synthesized DNA would be labeled with a stronger label and appear darker on the images.
- Visualization of chromosomes revealed darker segments on BOTH ends of the replication bubble.
Conclusion: Replication indeed begins at one location of origin of replication, generating a replication bubble, and DNA at both ends of the bubble unwinds in opposite directions, replicating DNA both ways away from the origin at the replication forks.
5.2.4 DNA Polymerase Catalyzes Phosphodiester-Linkage Formation
Before we consider what happens at replication forks in detail, let’s focus our attention on DNA POLYMERASES- a class of enzymes that catalyze the step-wise addition of nucleotides to a DNA strand.
DNA Polymerase enzymes have some unique properties:
- DNA polymerase is a template-directed enzyme that synthesizes a product with a base sequence complementary to that of the template.
BUT
- All DNA polymerases discovered to date can only elongate a preexisting DNA or RNA strand, they cannot initiate chains!
How would DNA synthesis begin then?
Solution: RNA polymerases do not require a preexisting base-paired 3′ end to initiate synthesis.
In the cell, an RNA polymerase enzyme, called Primase assembles a short stretch of RNA – the PRIMER base paired to the parental DNA template. Given the similarity between RNA and DNA, DNA polymerases can extend the free 3′ OH group provided by RNA!
The PRIMER-TEMPLATE junction is the cellular substrate recognized by DNA polymerase.
Primase is a slow (relatively!) and error-prone polymerase. The error-prone nature of its activity is not a problem, because the cell will remove all the RNA primers later in the process of DNA synthesis and replace them with DNA nucleotides- and we’ll revisit this later in this reading.
Chemistry of DNA Synthesis
Synthesis of DNA involves adding nucleotides, one by one, in the exact order specified by the original (template) strand.
DNA polymerase catalyzes the reaction by which an incoming deoxyribonucleotide, complementary to the template, is added onto the 3′ end of the previous nucleotide. The importance of the 3’OH group lies in the nature of the reaction that builds a chain of nucleotides.
The reaction catalyzed by the DNA polymerase occurs through the nucleophilic attack by the 3’OH group of the last nucleotide of the growing strand on the α phosphate of the incoming dNTP (Figure 5.4). The 5′ phosphate of the new nucleotide binds to the 3′ OH group of the nucleotide to make a phosphodiester bond.
The immediate hydrolysis of the pyrophosphate that is cleaved off the incoming dNTP drives the reaction forward.
Each added nucleotide provides a new 3’OH, allowing the chain to be extended for as long as the DNA polymerase continues to synthesize the new strand.
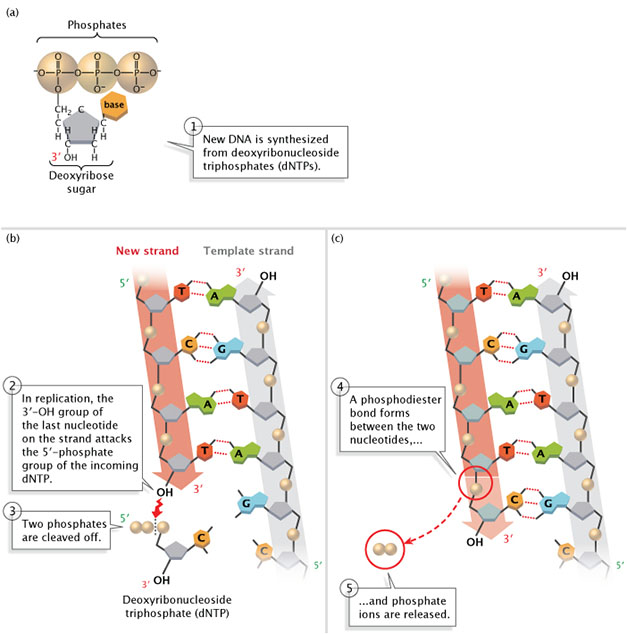
Thus DNA polymerases can only extend a strand in the 5′ to 3′ direction. Scientists have yet to identify a polymerase that can add bases to the 5′ ends of DNA strands.
Now consider one replication fork. Here the DNA strands of separated, and the DNA ahead of the fork remains wound in a double helix and the separated strands serve as templates for new daughter strand synthesis.
Remember this fork is moving- the downstream DNA is continuously being unwound and upstream DNA is continuously being replicated into hybrid helices of old and new DNA strands. But this DNA has to be anti-parallel to the parental DNA.
Time to Think
So if no 3’ to 5’ synthesizing activity can be found, how is the new strand oriented 3’ to 5’ in the direction of the replication fork synthesize without violating one of the two fundamental rules of nucleic acid chemistry?
5.2.5 DNA synthesis at the replication fork semi-discontinuous
This conundrum is solved, by copying DNA continuously on one template strand and discontinuously on the other.
One template strand is oriented such that the 5’-to-3’ direction in which the daughter strand synthesis is taking place is in the direction of fork movement; this strand can be synthesized in a CONTINUOUS manner and is known as LEADING STRAND synthesis.
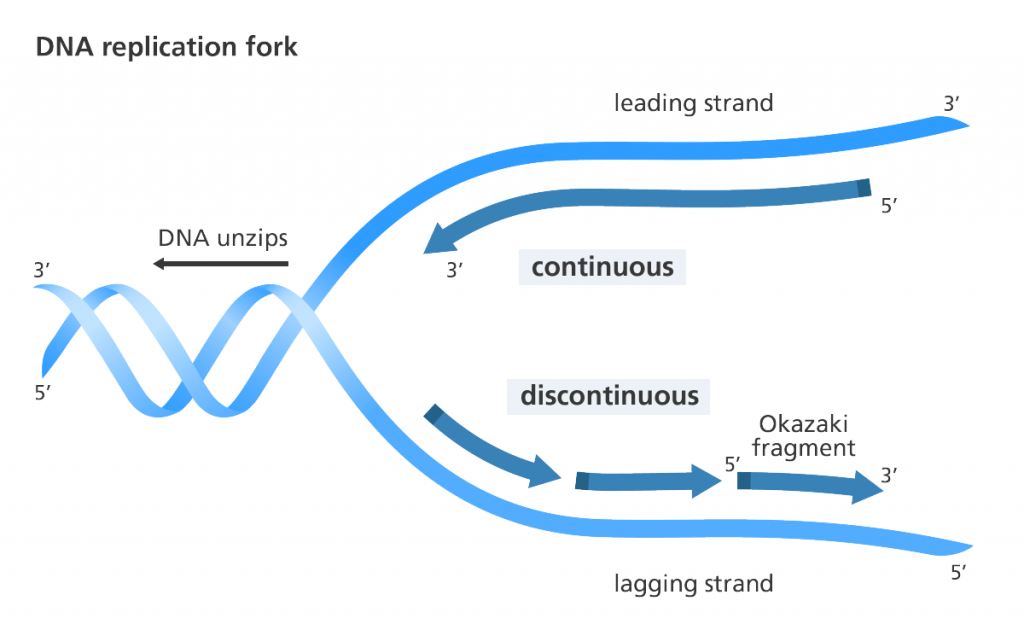
DNA copied from the other template strand, on the other hand, is made in short segments away from the fork or discontinuously.
This is known as LAGGING STRAND synthesis. Lagging strand synthesis takes place in a 5’-to-3’ direction and no rules of nucleic chemistry are violated.
Eventually, these short segments of DNA will have to be stitched together by an enzyme called DNA LIGASE.
The short stretches of DNA generated during lagging strand synthesis are commonly known as Okazaki fragments after their discoverer Reiji Okazaki. In bacteria, Okazaki fragments are 1,000-2,000 nucleotides in length, whereas in the cells of higher organisms they are typically only 100-200 nucleotides long.
See here for an animation of this process: Animation of Bidirectional Replication (YouTube)
Did I Get This?
Key Takeaways
- Replication is initiated at origin, creating two replication forks that move bidirectionally (in opposite directions)
- All DNA polymerases require a 3′-OH end to initiate DNA synthesis.
- The DNA polymerase advances continuously when it synthesizes the leading strand, but synthesizes the lagging strand in short fragments (Okazaki fragments)
Study Tip. Pause here and watch the Lecture Video: Chemistry of DNA Synthesis
Prof Mehta’s Study Note: An Important Distinction:
Bidirectional replication means that there are 2 replications forks moving in opposite directions. It does NOT refer to the fact that the 2 DNA strands (leading and lagging strands) are made in opposite directions. That is called semi-discontinuous synthesis. At every fork DNA replication is semi-discontinuous.
5.3 Structure of DNA Polymerase
The three-dimensional structures of a number of DNA
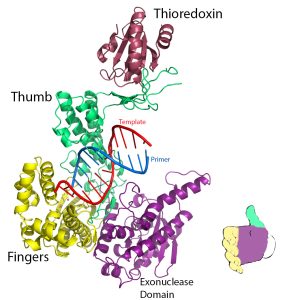
polymerase enzymes are known. The shape has been compared to that of a hand (or baseball mitt) with the protein domains that are referred to as the fingers, the thumb, and the palm. (see diagram )
Each subdomain carries a specific function. The fingers domain binds the incoming dNTP, the thumb domain helps grip duplex DNA, and the palm domain contains the amino acids that are part of the active site of the enzyme. The catalytic palm domain is the most conserved of the domains,
5.3.1 Accuracy of Replication
Changes in DNA sequence (mutations) can change the amino acid sequence of the encoded proteins and can often (though not always!) be harmful to the functioning of the organism.
When billions of bases in DNA are copied during replication, how do cells ensure that the newly synthesized DNA is a faithful copy of the original information?
DNA synthesis is extremely accurate with only one error per 109 bases!
3 Mechanisms contribute to the remarkable accuracy of replication
- The change in the structure of the enzyme when it binds the correct nucleotide. The DNA polymerase clasps the ds DNA tightly before binding the new dNTP. Within the active site of a DNA polymerase, the incoming dNTP is tested. A perfect fit triggers a conformational change: the finger domain rotates to form a tight pocket into which only a properly shaped base pair will readily fit.
Recall that the complementary base pairing (A with T and G with C) results in just the correct width for DNA!
The proper base is favored by the formation of a base pair, which is stabilized by specific hydrogen bonds. The binding of a non-complementary base is less likely because the interactions are energetically weaker.
2. 3′-5′ Exonucleolytic Proofreading: DNA polymerases are their own editors! They can correct mistakes in DNA by removing mismatched nucleotides. This happens with a separate 3′-5′ EXONUCLEASE activity that allows it to snip out the incorrect base and replace it with the correct base and resume replicating the template strand.
3. Strand Directed Mismatch Repair: A special mechanism inside cells to correct mismatches that already got incorporated.
We will learn about mismatch repair in the DNA Repair unit.
Animation: https://youtu.be/6hcK–4S68U
Before you continue you should
Complete any exercises and watch associated lecture videos
5.4 Process of Replication
Our understanding of the process of DNA replication is derived from studies using bacteria, yeast, and other systems. These investigations have revealed that DNA replication is carried out by the action of a large number of proteins that act together as a complex protein machine. (Table 5.1) This complex machine is known as REPLISOME.
Although the specific proteins involved are different in bacteria and eukaryotes, the basic mechanisms and principles are relevant in all cells.
We will primarily describe the process as discovered in Prokaryotes and point out differences with Eukaryotes towards the end.
5.4.1 Stages of Replication
As with many processes in molecular biology, we conventionally look at replication as being made up of three phases – initiation, elongation, and termination.
| Stage | What happens | Goal |
| INITIATION: Getting Started | DNA first unwound at the origins of replication. | Make space!! Open up the ds DNA to allow the machinery to load!!
Assembly of the replication apparatus on the origin of replication. |
| ELONGATION: Making DNA | DNA is made!
The leading strand is synthesized continuously The lagging strand is synthesized discontinuously. |
Make DNA, avoid errors, and be efficient (fast!)
|
| TERMINATION: Stop | 2 replication forks meet about halfway around the bacterial chromosome. | Avoid re-replication. Ensure completion of replication.
Disengage apparatus and proteins. |
5.4.1 Stage 1: Initiation of Replication
As we have seen, DNA synthesis starts at one or more origins or replication.
How does the replication machinery know where to begin?
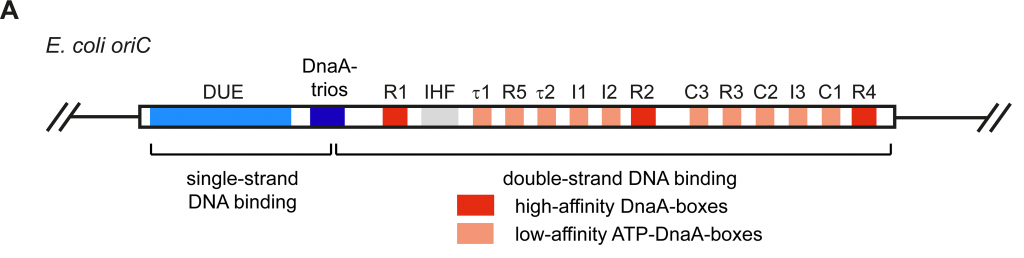
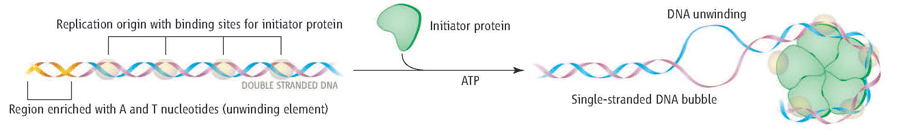
Origins are specific nucleotide sequences where replication begins. In E. coli (as do most prokaryotes), there is a single origin of replication (ori-C) on its one chromosome. This region is depicted in Figure 5.6
https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1008320
The region includes:
- Several conserved sequences which serve as binding sites for a protein called DnaA [DnaA boxes]
- An AT-rich region also called the DNA Unwinding Element
- GATC Methylation sites. (not shown in Figure above)
A-T base pairs have two hydrogen bonds between them and are more readily disrupted than G-C base pairs which have three. Therefore DNA with sequence of repetitive A-Ts can easily ‘open up’.
Initiation of replication begins when INITIATOR PROTEINS (DnaA for E. coli) will recognize and bind to the sequences present within the origins of replication.
For E.coli
- DnaA proteins bind the DnaA- Boxes (orange above)- these are the recognition sequences for the binding of the DnaA initiator protein.
- The binding of these proteins promotes the recruitment of multiple DnaA subunits that form a helical oligomer
- Bending of the DNA and torsional stress promotes the melting of the adjacent region (AT)
Unwinding
Once a small region of the DNA is opened up at each origin of replication, the DNA helix must be unwound to allow replication to proceed.
How are the strands of the parental DNA double helix separated?
The unwinding of the DNA helix requires the action of a class of enzymes called HELICASES.
Helicases are motor proteins that move (at speeds up to that of a jet engine!) directionally along a nucleic acid phosphodiester backbone, separating two annealed nucleic acid strands such as DNA and RNA, using energy from ATP hydrolysis.
Helicase loads onto the lagging strand and is a ring-shaped protein.
Note that a replication bubble is made up of two replication forks that “move” or open up, in opposite directions. At each replication fork, the parental DNA strands must be unwound to expose new sections of the single-stranded template.
[In E.coli the loading of the helicase is actually a two-step process but let’s not complicate things!]
Two problems arise as a result of unwinding DNA
- The separated single strands of DNA must be kept from coming back together so the new strands can continue to be synthesized.
Solution: To ensure that unwound regions of the parental DNA remain single-stranded and available for copying, the separated strands of the parental DNA are quickly coated by many molecules of a protein called single-strand DNA binding protein. (SSB)
2. A knotty problem: What is the effect of unwinding one region of the double helix? The local unwinding of the double helix causes over-winding (increased positive supercoiling) ahead of the unwound region. The DNA ahead of the replication fork has to rotate, or it will get twisted on itself and halt replication. This is a major problem, not only for circular bacterial chromosomes but also for linear eukaryotic chromosomes, which, in principle, could rotate to relieve the stress caused by the increased supercoiling.
Solution: Topoisomerases
Topoisomerases are special enzymes that can relieve the topological stress caused by local “unwinding” of the extra winds of the double helix. They do this by cutting one or both strands of the DNA and allowing the strands to swivel around each other to release the tension before rejoining the ends.
In E. coli, the topoisomerase that performs this function is called GYRASE.
DNA gyrase moves ahead of the replication fork and is essential for the process of replication.
Watch this animation to better understand the concept of supercoiling
Loading Primase
Okay, so we have opened up DNA. What’s next?
Remember DNA polymerases cannot begin synthesis of a new DNA strand de novo and require a free 3′ OH to which they can add DNA nucleotides.
The helicase recruits primase which synthesizes RNA primers used to initiate DNA synthesis.
Did I get this?
Key Takeaways
- The origin of replication in E.coli (oriC) contains conserved sequences called DnaA boxes and AT-rich regions.
- Initiator proteins (DnaA) bind to DnaA boxes, forming a complex that leads to the melting of DNA.
- DNA helicase forms at the replication fork.
- SSB – single-stranded binding proteins and DNA gyrase are also needed at the origin,
- Primase is bound to helicase and when activated synthesizes the RNA primer.
5.4.2 DNA Polymerases
E. coli has a total of five DNA polymerases. Three of these enzymes are involved in DNA replication (DNA polymerases I, II, and III). DNA polymerase III is the main polymerase involved in both leading strand biosynthesis and the synthesis of the Okazaki Fragments during DNA replication.
DNA polymerase III consists of 10 different proteins organized into three functionally distinct, but physically interconnected assemblies and referred to as DNA Pol-III Holoenzyme. (Figure 5.7)
A cartoon of the arrangement is shown on the right in the diagram below.
There are copies [the 3 glove-like structures (in black) in the cartoon] of DNA Pol-III catalytic core or enzyme. This is the protein responsible for synthesizing DNA. These are connected via flexible linker proteins to another sub-structure (green and red) that consists of several proteins that together make up the Sliding Clamp loader and Sliding clamp.
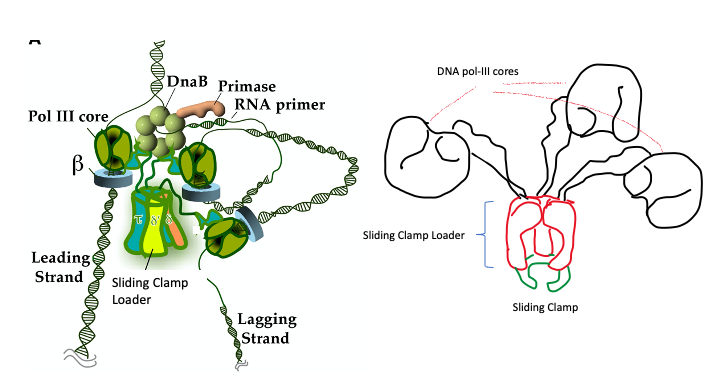
On right is a cartoon diagram of the DNA Pol III holoenzyme- illustration by S. Mehta.
Role of Sliding Clamp and Clamp Loader
Consider that the chromosome of E. coli consists of almost five million base pairs. Because DNA replication in E. coli takes place simultaneously from two replication forks, the overall rate of DNA synthesis is 1,600 nucleotides per second. Thus, E. coli is capable of duplicating its entire chromosome in as little as 40 minutes, and as we will see below, it does so with considerable accuracy.
This property of remaining attached to the DNA through many rounds of nucleotide addition is referred to as processivity.
Processivity maximizes the speed of DNA synthesis.
If the polymerase frequently fell off the DNA and had to re-bind in order to resume nucleotide incorporation, the rate of DNA synthesis would be much slower.
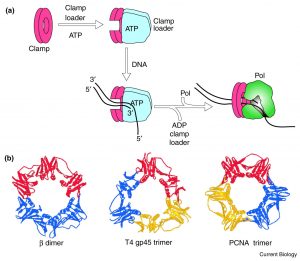
Volume 11 Issue 22 Pages R935-R946 (November 2001). Open Access DOI: https://doi.org/10.1016/S0960-9822(01)00559-0
This is where the sliding clamp and clamp loader comes in.
The shape of the clamp – a ring-shaped structure provides a geometrically elegant and simple solution.
DNA polymerase is tethered to the DNA by a sliding clamp, which (being ring-shaped) fully encircles the DNA helix.
Sliding clamps cannot load onto DNA spontaneously because they are closed circles. The job of the clamp loader is to ‘load’ the clamp onto the DNA.
Adenosine triphosphate (ATP)–dependent clamp loaders open the sliding clamps and load them onto primer-template junctions. Hydrolysis of ATP to ADP then changes the affinity of the clamp loader for the clamp and leaves it behind. The clamp spontaneously closes encircling the DNA helix. (Figure 5.8).
The sliding clamp is then joined by the DNA Polymerase. In the presence of the sliding clamp, DNA polymerases are much more processive, making replication faster and more efficient.
Note that the sliding clamp is loaded at the Primer-Template junction. Once the sliding clamp and DNA pol III cores are engaged the process of elongation begins!
Before you continue you should list/order the events that have taken place in Initiation thus far.
Let’s Review
5.4.3 Stage 2: Elongation of Replication
During elongation, the core polymerase (glove part of DNA pol III) adds DNA nucleotides to the 3′ end of the newly synthesized polynucleotide strand. The template strand specifies which of the four DNA nucleotides (A, T, C, or G) is added at each position along the new chain. Only the nucleotide complementary to the template nucleotide at that position is added to the new strand.
1. The leading and lagging strand polymerases are both tethered to the clamp loader. Recall, however, that the lagging strand (chemistry) necessitates synthesizing it in a direction opposite to the growing replication fork.
How can DNA be simultaneously synthesized on both the leading and lagging stands if the polymerases are bound to each other?
2. Note that there are 3 core polymerases. What is the purpose of the third polymerase?
In an updated model for replication known as the TROMBONE model of replication (see animation below). The lagging template strand is looped out so that it passes through the polymerase site in one subunit of polymerase III in the 3′ to 5′ direction.
After adding about 1000 nucleotides, DNA polymerase III lets go of the lagging-strand template by releasing the sliding clamp.
This mode of replication has been termed the trombone model because the size of the loop lengthens and shortens like the slide on a trombone!
This looping allows the primase and the Pol III active site can accomplish the discontinuous synthesis of the lagging template strand even though the general direction of the Pol III complex is opposite to the required direction of DNA synthesis.
Thus the lagging strand requires 2- Core polymerases (in E.coli) to allow for replication to be completed in a timely fashion. One core-polymerase engages with the looped Okazaki fragment. It begins extension and the DNA is released as helicase proceeds forward.
As the core polymerase continues to extend the Okazaki fragment a newly primed section is formed at the replication fork, which is then captured by the third polymerase.
DNA Replication Animation below highlights the whole process:
Completing Elongation
As each newly formed segment of the lagging strand approaches the 5′ ends of the adjacent Okazaki fragment (the one just completed).
How are these RNA/DNA fragments converted into one long continuous DNA strand?
The RNA could be removed by a polymerase that has 5′->3′ exonuclease activity, however, Pol III lacks this activity.
Unlike polymerase III, DNA polymerase I has a 5′ → 3′ exonuclease activity.
In E.coli,
- RNAse H first removes all the ribonucleotides in the primer except for the rNTP linked to the DNA end.
- DNA Pol I use nick-translation- utilizing its polymerizing ability to extend the 3′ end of the newly formed Okazaki fragment and the 5′-3′ exonuclease activity to remove the remaining ribonucleotide from the neighboring Okazaki fragment.

Finally, another critical enzyme, DNA ligase, joins adjacent completed fragments, making the final phospho-diester bond in the ‘nicks’ left behind between Okazaki fragments. (Figure 5.9)
5.4.4 Stage 3: Termination of Replication
Proper termination of DNA replication is important for genome stability. E. coli replication terminates in the region opposite oriC.
The two replication forks meet at a termination region, setting off a series of events that leads to the eventual completion of replication and subsequent chromosome separation.
The termination region contains special conserved sequences called (Ter) sites. These sequences are recognized by Tus proteins.
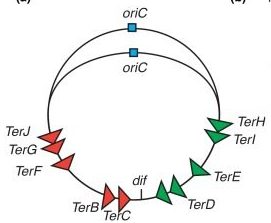
The 10 Ter sites are split and organized as two oppositely orientated groups of five. The association of the Tus proteins to the Ter sites is also polar, such that there is a permissive face that allows the replisome to pass unhindered and a non-permissive face that can block the replisome (Figure 5.9).
This configuration allows the replisome to pass the first Tus-Ter complex unhindered and be blocked at the second. When the 2 replisomes meet the apparatus disassembles.
We now have 2 circular chromosomes that are linked to one another. Here the activity of a different enzyme from the Topisomerase family becomes useful in separating or un-linking the 2 chromosomes.
Practice: Terminology Crossword
Before you continue you should
- Watch the Lecture videos that cover the material above (if you haven’t)
5.5 Eukaryotic Replication
Replication in eukaryotes while mechanistically similar to replication in bacteria is more challenging in the following ways.
1) Size: E. coli must replicate 4.6 million base pairs, whereas a human diploid cell must replicate 6 billion base pairs.
2) The second challenge is the fact that while the genetic information for E. coli is contained on one chromosome, human beings have 23 pairs of chromosomes that must be replicated.
3) While the E. coli chromosome is circular, human chromosomes are linear. The third challenge arises because of the nature of DNA synthesis on the lagging strand. Linear chromosomes are subject to shortening with each round of replication unless countermeasures are taken.
4) The events of replication have an additional twist in eukaryotes. Recall that DNA is found in eukaryotic cells as chromatin, a complex of the DNA with proteins. The nucleosome structure must be disrupted to make DNA available for replication and restored after replication is completed.
The solution to Challenge 1 and 2 :
Origin organization, specification, and activation in eukaryotes are more complex than in bacterial or archaeal kingdoms and significantly deviate from the paradigm established for prokaryotic replication initiation.
To account for replicating large genome sizes – eukaryotic chromosomes have multiple origins of replication, which are located between 30 and 300 kilobase pairs (kbp) apart.
In human beings, replication of the entire genome requires about 30,000 origins of replication, with each chromosome containing several hundred.
Each origin of replication represents a replication unit or replicon, and at each origin, replication is semi-discontinuous and bi-directional.
The essential steps of replication are the same as in prokaryotes:
Before replication can start, the DNA has to be made available as a template.
– A helicase using the energy from ATP hydrolysis opens up the DNA helix.
– Replication forks are formed at each replication origin as the DNA unwinds.
– The opening of the double helix causes over-winding, or supercoiling, in the DNA ahead of the replication fork. These are resolved with the action of topoisomerases.
– Primers are formed.
The differences often are in the number of proteins needed for these functions.
For example:
1. The protein needed to bind the sequences that define the origins in eukaryotic genomes is part of a large complex of proteins- Origin Recognition Complex.
2. The number of DNA polymerases in eukaryotes is more than those found in prokaryotes: 14 are known, of which five are known to have major roles during replication and have been well studied. They are known as pol α, pol β, pol γ, pol δ, and pol ε.
At the eukaryotic replication fork, three distinct replicative polymerase complexes contribute to canonical DNA replication: α, δ, and ε.
The ‘Primase’ for eukaryotes is DNA polymerase α (Pol α) – This protein has 2 parts- one which synthesizes DNA (not unlike all the DNA polymerases) the other has RNA priming activity.
Thus this complex accomplishes the priming task by synthesizing a primer that contains a short ~10-nucleotide RNA stretch followed by 10 to 20 DNA bases.
Importantly, this priming action occurs at origins to begin leading-strand synthesis and also at the 5′ end of each Okazaki fragment on the lagging strand.
However Pol α is not able to continue DNA replication- it is not processive!
3. After priming, synthesis is “handed off” to 2 other polymerases to continue elongation. The polymerase switching requires clamp loaders. The leading strand is ‘handed off’ to the enzyme pol δ, the lagging strand to pol ε.
5.5.1 Dealing with Histones
As seen in Chapter 4 chromosomes are packaged by wrapping ~147 nucleotides (at intervals averaging 200 nucleotides) around an octamer of histone proteins, forming the nucleosome. The histone octamer includes two copies each of histone H2A, H2B, H3, and H4.
It was highlighted that histone proteins are subject to a variety of post-translational modifications, including phosphorylation, acetylation, methylation, and ubiquitination that represent vital epigenetic marks.
The nucleosome structure must be disrupted to make DNA available for replication and restored after replication is completed.
Furthermore, it is important to transmit the epigenetic information found on the parental nucleosomes to the daughter nucleosomes, in order to preserve the same chromatin state.
In other words, the same histone modifications should be present on the daughter nucleosomes as were on the parental nucleosomes. This must all be done while doubling the amount of chromatin, which requires the incorporation of newly synthesized histone proteins.
This process is accomplished by histone chaperones and chromatin remodeling complexes.
Ahead of the replication fork, the chromatin structure is disassembled by ATP-dependent chromatin remodeling complexes, allowing access to the DNA template. The loss of the histone octamer from the parental DNA during DNA replication is accompanied by the dissociation of H3/H4 tetramers and H2A/H2B dimers.
Special histone protein chaperones recruit histone H3-H4 dimers to the replication fork- helping load both newly synthesized (light purple) histones to establish chromatin behind the fork. Previously loaded histones (dark purple) are also deposited on both daughter DNA strands.
Thus in the reassembled chromatin, half of the histones are recycled from the parental chromatin while the other half are newly-synthesized.
(Figure 5.12)
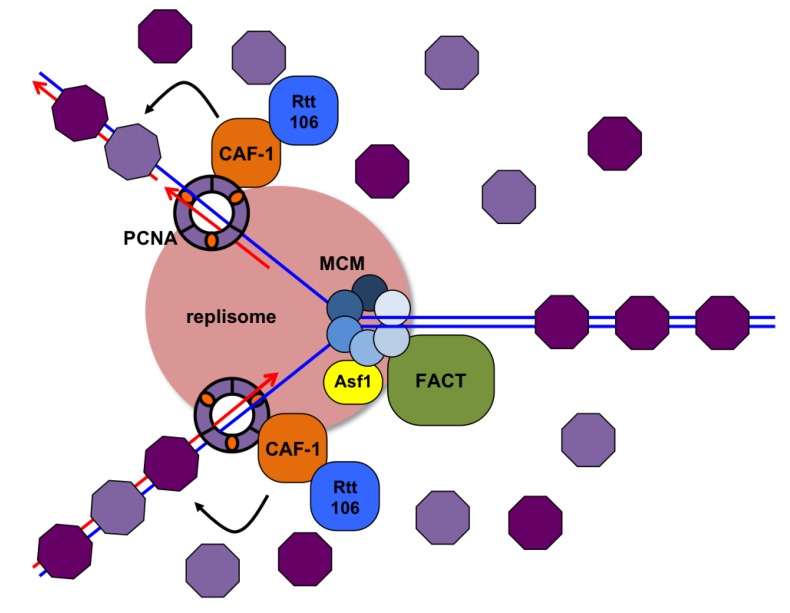
The presence of some recycled histones allows for one possible mechanism of how the epigenetic state is re-established. Presumably, the old histones would still be carrying the modifications (acetyl or methyl groups for example) which can then facilitate the spread of modifications using Bromo and Chromodomain-containing complexes that have histone-modifying activity.
For example, old histone bearing an acetyl group is recognized by a bromodomain-containing Histone acetyl Transferase, which will then acetylate adjacent histones.
Scientists are still uncovering how this important aspect of ‘somatic’ cell identity is maintained.
5.5.1 End Replication Problem- Replicating Telomeres
As you’ve learned, the enzyme DNA polymerase can add nucleotides only in the 5′ to 3′ direction. Leading strand synthesis continues until the end of the chromosome is reached. On the lagging strand, DNA is synthesized in short stretches, each of which is initiated by a separate primer.
When the replication fork reaches the end of the linear chromosome, there is no way to replace the primer on the 5’ end of the lagging strand leaving a structure called a 3’-overhang or a 5’-gap (Fig 5.13).
Interestingly, this overhang is critically important in forming a ‘cap-like structure at the end of the chromosome called the telomere.
The telomere has a highly repetitive noncoding sequence that is recognized by proteins that bind the DNA sequence and ‘hide’ the linear end of the chromosome by forming a loop. This structure is important because it is telling the cell “Hey, I’m the normal end of the chromosome-leave me alone; don’t try and fix me!”
Because the cell needs this 3’-overhang to create the correct end structure, the leading strand is resected (or chewed up) by an exonuclease to create a 3’-overhang.
After each round of replication, the leading strands are continually shortened (the lagging strands also gradually shorten due to the removal of the last RNA primer, but this shortening is actually less detrimental compared to the degradation on the leading strands). If the shortening at the ends of the chromosomes was not fixed, eventually the highly repetitive sequence of the telomere would be lost and the cap structure at the ends of the chromosomes would no longer form, causing the cell to signal that the DNA was damaged.
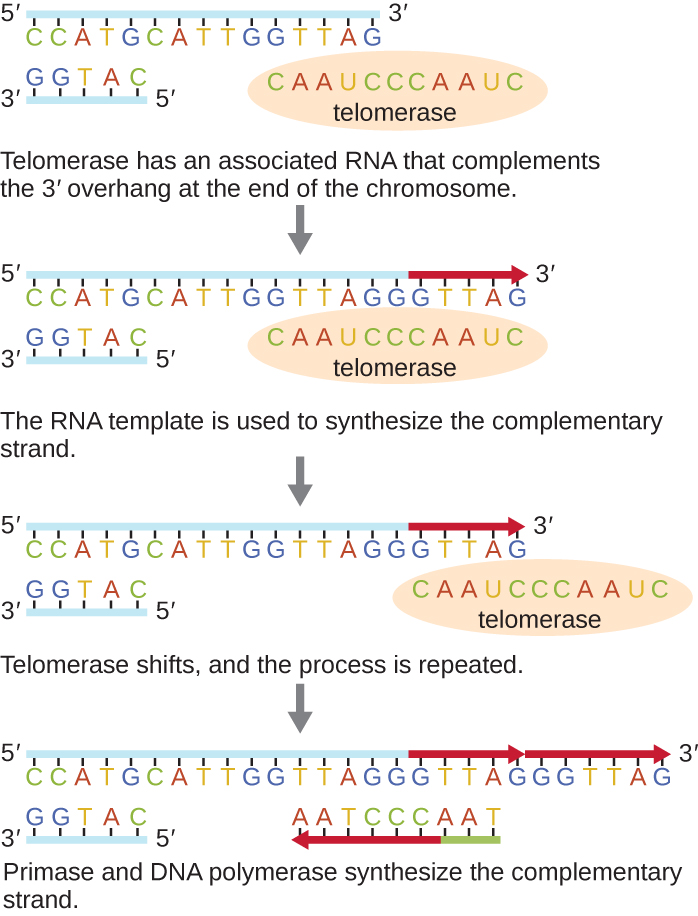
To combat the loss of DNA sequence in the telomere and prevent a DNA damage response, an enzyme called telomerase (Figure 5.13), adds nucleotides to the ends of chromosomes.
Telomerase is highly expressed in early development and gametic cells; it is moderately expressed in many adult stem cells, but not expressed in most adult somatic cells.
Telomerase is an enzyme that contains a catalytic part and a built-in RNA template. Because it uses an RNA template to build DNA (transcription in reverse), it is called reverse transcriptase.
Telomerase attaches to the 3’-overhang (location of a 3’OH group) of the chromosome and adds DNA nucleotides that are complementary to the RNA template.
Once the 3′ end of the template is sufficiently elongated, DNA polymerase can add a new primer and fill in the nucleotides complementary to the ends of the chromosomes (Figure 5.13).
The final RNA primer will be removed, re-establishing the 3’-overhang and allowing the cap structure to be formed once again.
In many cancer cell types, the expression of the telomerase activity has been reactivated, allowing cancer cells to lengthen telomeres and become immortal.
Because of this action, there is great interest in better understanding how telomerase and the telomere are regulated in cells and if there are ways to exogenously control the enzyme.
Link to Learning
This You-Tube Video compares the process of prokaryotic and eukaryotic replication
Concepts in Context: Telomeres and Ageing.
Cells that undergo cell division continue to have their telomeres shortened because most somatic cells do not make telomerase. This essentially means that telomere shortening is associated with aging. With the advent of modern medicine, preventative health care, and healthier lifestyles, the human life span has increased, and there is an increasing demand for people to look younger and have a better quality of life as they grow older.
For her discovery of telomerase and its action, Elizabeth Blackburn (1948–) received the Nobel Prize for Medicine or Physiology in 2009.
WATCH: TED Talk from Elizabeth Blackburn, where she highlights the discovery and data on the relationship between stress and telomerase.
COMPLETE: Don’t forget to complete the assignments associated with this video in CANVAS.
Check your understanding
References and Attributions
This chapter contains material taken from the following CC-licensed content. Changes include rewording, removing paragraphs and replacing with original material, and combining material from the sources.
- Bergtrom, Gerald, “Cell and Molecular Biology 4e: What We Know and How We Found Out” (2020). Cell and Molecular Biology 4e: What We Know and How We Found Out – All Versions. 13.
https://dc.uwm.edu/biosci_facbooks_bergtrom/13 - Works contributed to LibreTexts by Kevin Ahern and Indira Rajagopal. LibreTexts content is licensed by CC BY-NC-SA 3.0
- Fundamentals of Cell Biology by Shoshana D. Katzman, et al., Georgia Gwinnet College, licensed by Attribution-NonCommercial-ShareAlike 4.0 (available https://alg.manifoldapp.org/projects/fundamentals-of-cell-biology)
Figure image attributions are provided within the text.
D. Samson, L. H. John F. Cairns (1922–2018). Nat Struct Mol Biol 26, 149–150 (2019). https://doi.org/10.1038/s41594-019-0194-1. See here: https://rdcu.be/crdJs

