Molecular Biology: From DNA to RNA to Protein
6 Transcription in Prokaryotes
6.1 Introduction
In the preceding sections, we have discussed the replication of the cell’s DNA and the mechanisms by which the integrity of the genetic information is carefully maintained.
What do cells do with this information? How does the sequence in DNA control what happens in a cell?
If DNA is a giant instruction book containing all of the cell’s “knowledge” that is copied and passed down from generation to generation, what are the instructions for? And how do cells use these instructions to make what they need?
Genes must be expressed
In earlier chapters and from previous classes you have learned that all living organisms have genes, these genes carry information in the form of a code (temporary instructions or mRNA) that is used to make proteins. The language of the genome is universal. This is what allows bacteria to make human insulin!
This description of the flow of information from DNA to RNA to protein is often called the central dogma of molecular biology and is a good starting point for an examination of how cells use the information in DNA.
Recall that proteins are polymers, or chains, of many amino acid building blocks. The sequence of bases in a gene (that is, its sequence of A, T, C, G nucleotides) translates to an amino acid sequence.
A triplet is a section of three DNA bases in a row that codes for a specific amino acid.
Similar to the way in which the three-letter code d-o-g signals the image of a dog, the three-letter DNA base code signals the use of particular amino acid.
For example, the DNA triplet CAC (cytosine, adenine, and cytosine) specifies the amino acid valine. Therefore, a gene, which is composed of multiple triplets in a unique sequence, provides the code to build an entire protein, with multiple amino acids in the proper sequence (Figure 6.1).
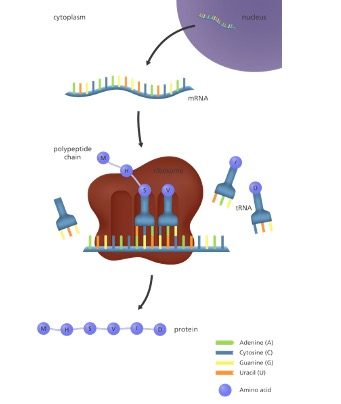
Because proteins are coded by genes, the term “gene expression” refers to protein synthesis (i.e., making proteins), including the regulation of that synthesis.
Gene Expression Is Regulated
Consider that all of the cells in a multicellular organism have arisen by division from a single fertilized egg and therefore, all have the same DNA. Division of that original fertilized egg produces, in the case of humans, over a trillion cells, by the time a baby is produced from that egg (that’s a lot of DNA replication!).
Yet, we also know that a baby is not a giant ball of a trillion identical cells, but has the many different kinds of cells that make up tissues like skin and muscle and bone and nerves.
How did cells that have identical DNA turn out so different? The answer lies in the REGULATION of GENE EXPRESSION which is the process by which the information in DNA is used. Although all the cells in a baby have the same DNA, each different cell type uses a different subset of the genes in that DNA to direct the synthesis of a distinctive set of RNAs and proteins.
We began to see glimpses of this idea when we discussed chromatin regulation.
In this section and through the rest of the sections we will unravel all the ways in which our GENOME is actually put to work.
We begin by examining RNA synthesis in the simple prokaryote- E. Coli. Like many of the discussions prior, studies in this organism gave insight into the basic biochemical processes that hold true in Eukaryotes as well – just with added complexity.
Learning Objectives
Level 1 and Level 2 (Knowledge and Comprehension)
- Explain what is meant by functional RNA
- Draw, describe or identify key features within a transcription unit.
- Explain the role of Sigma factor in bacterial transcription.
- Describe the three steps of transcription initiation that occur before the elongation phase begins.
- Explain the molecular mechanism behind transition from closed-open complex during initiation—(isomerization)
- Understand the meaning of promoter consensus sequences.
- Understand the different associations between RNA polymerase and DNA during transcription stages.
- Explain how termination of bacterial transcription occurs
- Distinguish between the 2 types of terminators sequences
- Explain the differences and similarities between RNA polymerase and DNA polymerase.
⊕ Level Up (Application, Analysis, Synthesis)
- When given an illustration that shows: a portion of a gene undergoing transcription, the template and coding strands labeled, and a DNA sequence you should be able to-
- Indicate the direction in which RNA polymerase moves as it transcribes this gene.
- Write the polarity and sequence of the RNA transcript from the DNA sequence given.
- Identify the location of the promoter for the gene.
- Identify elements of a gene, when given a gene sequence from a database.
- Predict how mutations in promoter sequences, genes coding for sigma factors would affect transcription.
6.2 Basics of Transcription
Transcription is the first step of the “central dogma” of information transfer from DNA to protein in which genetic information in genes is transcribed into RNA.
You can think of mRNA as being a copy of one book/ page within the book with the information needed for a particular assignment. While your genome is the entire library/master copy of the entire book that is in the restricted section of the library.
You can’t take that book out of the restricted section, so you need to make a copy of the information.

Are all the RNA’s in a cell messenger RNA (mRNA) molecules?
No! While we focus mainly on mRNA or ‘protein coding’ RNA when we think about transcription, less than 5% of total RNA in a cell is actually messenger RNA.
There are many other types of RNAs in the cell. These RNAs are referred to as functional or non-coding RNA (ncRNA). (Figure 6.2)
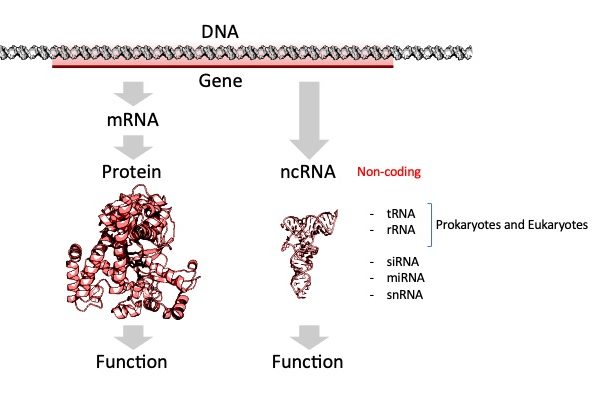
The RNA itself fulfills an important function, either enzymatic, structural, or as we shall see later in the semester in control of gene expression.
All cells (prokaryotes and eukaryotes) make three main kinds of RNA: ribosomal RNA (rRNA), transfer RNA (tRNA), and messenger RNA (mRNA).
tRNA carries the appropriate amino acids into the ribosome for inclusion in the new protein. Meanwhile, the ribosomes themselves consist largely of ribosomal RNA (rRNA) molecules. Quantitatively, rRNAs are by far the most abundant RNAs in the cell.
Other small ncRNAs and long ncRNA molecules play a role in the regulation of transcriptional and translational processes and we will learn about those in later chapters.
The diagram below shows the central dogma in prokaryotes and eukaryotes. See if you can spot the differences between them as it relates to transcription by comparing the diagrams below!
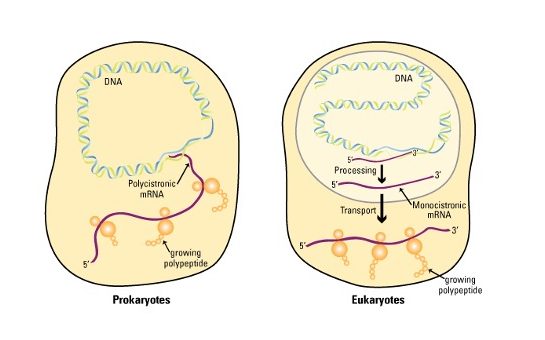
Location
Most RNA transcripts in prokaryotes emerge from transcription ready to use! In fact, transcription and translation are coupled, with the association of ribosomes with mRNA and the translation of a polypeptide beginning even before the transcript is finished. This is because these cells have no nucleus.
Eukaryotic transcripts must exit the nucleus before they encounter the ribosomes in the cytoplasm.
mRNA type :
Eukaryotic transcripts have to undergo additional processing by trimming, splicing, or both!
In contrast to eukaryotes, some bacterial genes are part of operons whose mRNAs encode multiple polypeptides.
Chromatin:
DNA in bacteria is virtually ‘naked’ in the cytoplasm while eukaryotic DNA is wrapped up in chromatin proteins in a nucleus.
6.3 Rules of Transcription and Terminology
Since we are coming straight off a unit of Replication, it is useful in thinking about these 2 processes by comparing the 2 polymers (DNA v/s RNA) – we can use that fundamental information to get a basic outline of transcription and the rules of transcription like we did for replication.
Building an RNA strand is very similar to building a DNA strand. This is not surprising, knowing that DNA and RNA are very similar molecules.
Transcription is catalyzed by the enzyme RNA Polymerase. “RNA polymerase” is a general term for an enzyme that makes RNA. There are several different kinds of RNA polymerases in eukaryotic cells, while in prokaryotes, a single type of RNA polymerase is responsible for all transcription.
Unlike DNA replication, however, only short sections of the genome (the genes) are transcribed. Different genes may be copied into RNA at different times in the cell’s life cycle.
Further, our cells need billions of copies of proteins which will be made using these instructions. Therefore unlike DNA replication, many copies of RNA are made during transcription.
RNA synthesis: The substrates
Like DNA polymerases, RNA polymerases synthesize new strands only in the 5′ to 3′ direction, but because they are making RNA, they use ribonucleotides (i.e., RNA nucleotides) rather than deoxyribonucleotides. (Figure 6.4)
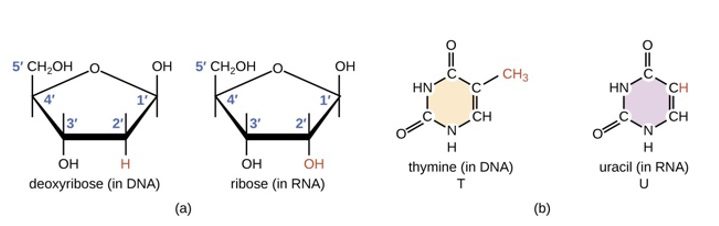
Ribonucleotides are joined in exactly the same way as deoxyribonucleotides, i.e., the 3’OH of the last nucleotide on the growing chain is joined to the 5′ phosphate on the incoming nucleotide to make a phosphodiester bond.
One important difference between DNA polymerases and RNA polymerases is that the latter does not require a primer to start making RNA.
Once RNA polymerases are in the right place to start copying DNA, they just begin making RNA by joining together RNA nucleotides complementary to the DNA template.
RNA synthesis: The Template
A gene (DNA) is double-stranded, but only one is transcribed into RNA!
Within each section of a ‘gene’, ONE of the TWO strands carries the code/information to make the protein and is referred to as the sense strand or coding strand. (Figure 6.5)
The goal of transcription is to make a copy of that code accurately such that the 5′ end of the code is also represented at the 5′ end of the RNA!
Therefore during transcription, the opposite strand (antisense) or non-coding strand is used as a template.
RNA that is synthesized is complementary and antiparallel to the DNA template strand.
We can differentiate the two strands of DNA on the basis of their relation to the RNA product.
The sequence of the template strand of DNA is the complement of that of the RNA transcript. It is also referred to as the non-coding strand or anti-sense (-) strand.
In contrast, the coding strand of DNA has the same sequence as that of the RNA transcript except for thymine (T) in place of uracil (U). The coding strand is also known as the sense (+) strand.
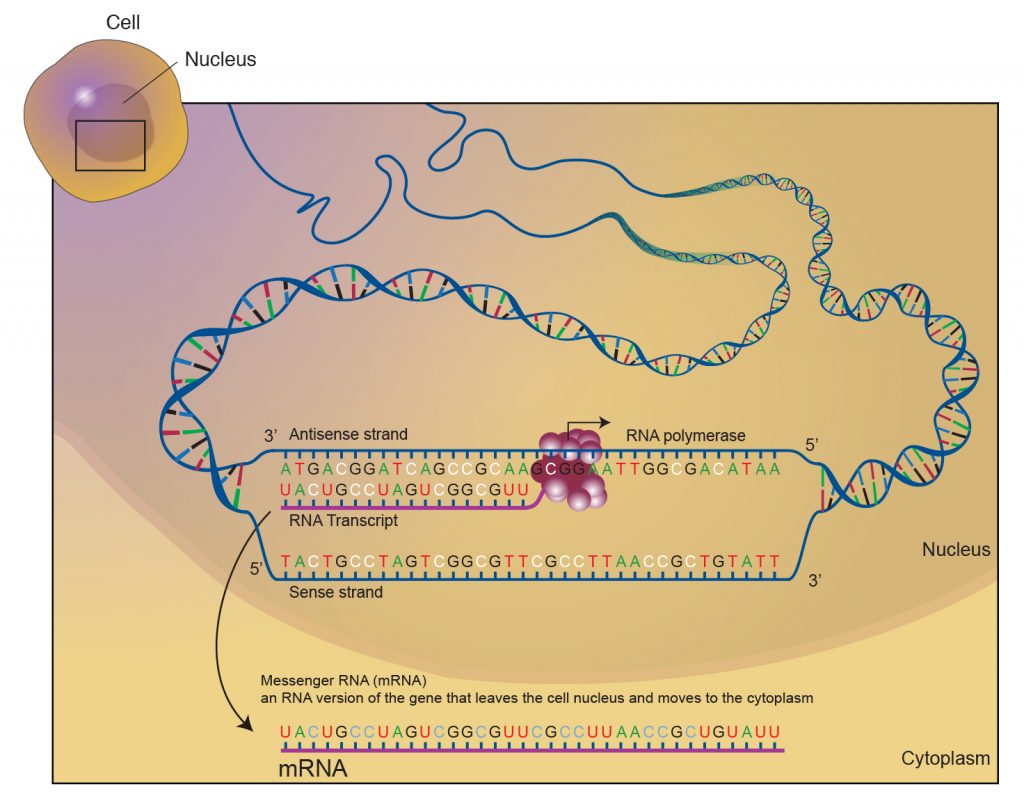
Either of the 2 polynucleotide strands may contain a gene, and hence the determination of sense and antisense is gene-specific!
(This is very important to understand!)
Did I Get This?
6.4 Genes are Transcription Units
Unlike the situation in replication, where every nucleotide of the parental DNA must eventually be copied, transcription, as we have already noted, only copies selected portions of the DNA into RNA at any given time.
Consider the challenge here: in a human cell, there are approximately 6 billion base pairs of DNA. Much of this is non-coding DNA, meaning that it will not need to be transcribed. The small percentage of the genome that is made up of coding sequences still amounts to between 20,000 and 30,000 genes in each cell. Of these genes, only a small number will need to be expressed at any given time.
This imposes a fundamental problem on the cell: how to recognize individual genes and transcribe them at the proper time and place. How do RNA polymerases know which DNA strand to read and where to start and stop?
That information is carried within the DNA sequence itself. There are patterns that indicate where RNA polymerase should start and end transcription.
The signatures within the DNA sequence also help a scientist (you!) take a sequence of letters and identify the beginning, middle, and end of the gene.
These sequences are recognized by the RNA polymerase or by proteins that help RNA polymerase determine where it should bind the DNA to start transcription.
Gene Structure:
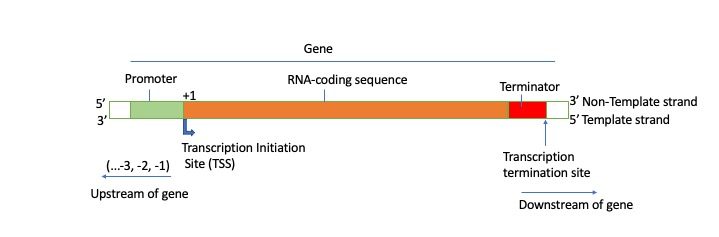
The stretch of DNA that encodes an RNA molecule and includes all the sequences necessary for its transcription is the transcription unit.
Much of the gene structure is broadly similar between eukaryotes and prokaryotes. These common elements largely result from the shared ancestry of cellular life in organisms with roughly 3.8 billion years of evolution.
Key differences in gene structure between eukaryotes and prokaryotes reflect their divergent transcription and translation machinery. Understanding gene structure is the foundation of understanding gene annotation, expression, and function.
Promoters:
Special DNA sequences, called promoters, direct the RNA polymerase to the proper site for the initiation of transcription.
A promoter is described as being situated upstream of the gene that it controls (Figure 7.57). What this means is that on the DNA strand that the gene is on, the promoter sequence is “before” the gene, or to put it differently, it is on the side of the gene opposite to the direction of transcription.
In this manner, the promoter actually indicates which of the two DNA strands is to be read as the template and the direction of transcription.
Also, notice that the promoter is said to “control” the gene it is associated with.
This is because the expression of the gene is dependent on the binding of RNA polymerase to the promoter sequence to begin transcription. If the RNA polymerase and its helper proteins do not bind at the promoter, the gene cannot be transcribed and it will, therefore, not be expressed. Promoters also control the frequency of transcription.
Coding Region (Open Reading Frame):
The first nucleotide that will be transcribed into RNA is the transcription start site and given the number +1. (Note: the promoter is NOT part of the mRNA!)
All nucleotides added after form the open reading frame or RNA -coding region.
Terminator Region:
The DNA sequence that indicates the endpoint of transcription, where the RNA polymerase should stop adding nucleotides and dissociate from the template is known as a terminator sequence.
Terminators are usually part of the RNA-coding sequence; transcription stops only after the terminator has been copied into RNA.
Upstream and Downstream:
When DNA sequences are written out, often the sequence of only one of the two strands is listed. Molecular biologists typically write the sequence of the nontemplate strand (coding strand) because it will be the same as the sequence of the RNA transcribed from the template strand.
Upstream refers to sequences in the opposite direction from expression and are assigned negative numbers.
Nucleotides downstream of the start site are assigned positive numbers. There is no nucleotide numbered 0.
Key Takeaways
- During Transcription, only certain sections of the DNA are transcribed at any one time.
- RNA is transcribed from a single strand of DNA. Within a gene, only one of the two DNA strands—the template strand—is usually copied into RNA.
- Ribonucleoside triphosphates are used as the substrates in RNA synthesis.
- The transcribed RNA molecule is antiparallel and complementary to the DNA template strand. Transcription is always in the 5′→3′ direction, meaning that the RNA molecule grows at the 3′ end.
- The transcription unit contains all of the sequences that are necessary for both making the RNA as well as regulating it.
- Upstream of the start of the gene are Promoter sequences that are crucial for the binding of RNA polymerase to DNA.
- Terminator sequences signal the end of the gene, these are transcribed and found in RNA.
6.5 RNA Polymerase Enzymes
RNA Polymerase Enzymes (RNAPs) are required to carry out the process of transcription and are found in all cells ranging from bacteria to humans. All RNAPs are multi-subunit assemblies (an example of a quaternary structure) and carry out the same reaction.
Bacterial RNAPs are the simplest form of RNA polymerases and provide an excellent system to study how they control transcription.
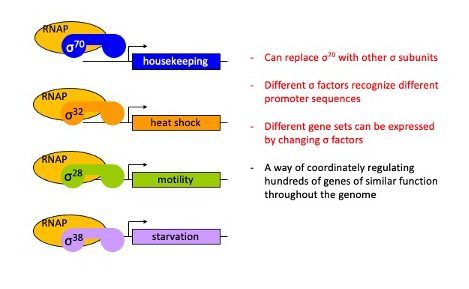
The RNAP catalytic core within bacteria contains five major subunits (α2ββ’ω- 2 copies of alpha, beta, beta prime and omega) (Fig 10.7B). To position this catalytic core onto the correct promoter requires the association of a sixth subunit called the sigma factor (σ).
Together, the σ subunit and core polymerase make up what is termed the RNA polymerase holoenzyme.
The core polymerase is the part of the RNA polymerase that is responsible for the actual synthesis of the RNA, while the σ subunit is necessary for binding the enzyme at promoters to initiate transcription. The sigma (σ) subunit helps to find a site where transcription begins, participates in the initiation of RNA synthesis, and then dissociates from the rest of the enzyme.
The first sigma factor to be identified was sigma70 (σ70) because it has a mass of 70kDa.
σ70 is the housekeeping sigma factor that is responsible for transcribing most genes in growing cells. It keeps essential genes and pathways operating.
How We Know
Biochemical assays combined with protein purification techniques of the kind you saw in Chapter 2 led to the identification of Bacterial RNA polymerase subunits.
It was found that RNA polymerase activity was associated with two protein species. A core polymerase (with subunit structure α2ββ′) can transcribe DNA into RNA inefficiently and nonspecifically. When the sigma subunit, σ70, is added, it can bind to core forming a holoenzyme (α2ββ′σ) that is capable of specific engagement with duplex DNA at the beginning of genes (promoters) as well as efficient initiation of transcription. (1)
WATCH Lecture Video 3 for the experiments that led to the identification and role of the sigma factor: L211 Introduction to Transcription (3): RNA polymerase
Before you continue you should
- Watch the Lecture videos that cover the material above.
- Complete the associated Lecture Quickcheck.
The role of the sigma factor is to help RNA polymerase find ‘authentic’ genes. Those with promoter sequences. What are those sequences?
Bacterial Promoters
Because the same RNA polymerase has to bind to many different promoters, it would be predicted that promoters would have some similarities in their sequences. Scientists examined many genes and their surrounding sequences and as expected, common sequence patterns were seen to be present in many promoters.
The locations of these nucleotides relative to the transcription start site are also similar in most promoters.
This common sequence pattern is called a CONSENSUS SEQUENCE. It is important to understand that each nucleotide in a consensus sequence is simply the one that appeared at that position in the majority (consensus) of promoters examined, and does not mean that the entire consensus sequence is found in all promoters.
The most common sequences found in bacteria are
A -10 sequence (Pribnow box): this is a 6 bp region centered about 10 bp upstream of the start site. The consensus sequence at this position is TATAAT. In other words, if you count back from the transcription start site, the sequence found at roughly -10 in the majority of promoters studied is TATAAT.
A -35 sequence: this is a 6 bp sequence at about 35 base pairs upstream from the start of transcription. The consensus sequence at this position is TTGACA.
The distance between these conserved sequences is 17-19bp. While the exact sequence of the spacer is not important that length with a separation of 17 nucleotides optimal.
The figure below shows the most common prokaryotic promoter: the σ70 promoter, so-called because it is recognized and bound by the σ70 transcription factor.
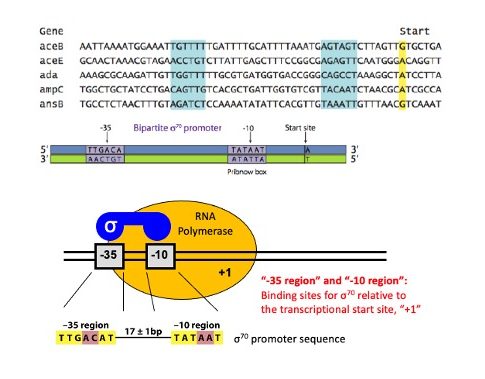
Notice the relationship between the various individual promoters and the consensus sequence. In general, those promoters with more matches to the consensus sequence are stronger promoters.
What does it mean to be a stronger (or weaker) promoter? First, keep in mind that the expression of any given gene is not automatic, or 100%. At any point in time, many of a cell’s genes will be near 0% or shut off.
However, even genes that are turned on are transcribed at different rates. One of the governing factors is the recognition of the promoter site.
Genes with strong promoters are transcribed frequently—as often as every 2 seconds in E. coli. The RNA polymerase holoenzyme (with sigma) is more likely to recognize the site, dock properly, open up the double helix, and begin transcribing.
In contrast, genes with very weak promoters are transcribed about once in 10 minutes. RNA polymerase can potentially recognize weaker promoters, but it is less likely to do so, instead of passing it by as just another unimportant stretch of DNA.
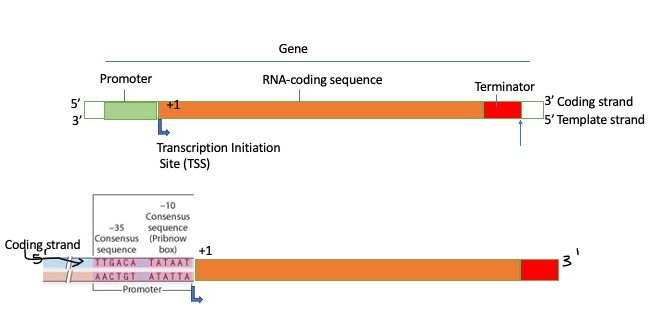
We can now add more detail to our transcription unit as shown below. Note that in the gene shown below the promoter is on the left of the start site.
Alternative Sigma Factors
Within bacteria, there are multiple different sigma factors that can associate with the catalytic core of RNAP that help to direct the catalytic core to the correct DNA locations where RNAP can then initiate transcription.
6.6 Steps in Transcription
Like DNA Replication, RNA polymerase synthesizes RNA in three distinct phases that are also conceptually similar
Initiation: ‘Setting up”. In this phase RNA polymerase holoenzyme locates and binds to promoter DNA to begin the synthesis of RNA>
Elongation: ‘The actual synthesis of RNA”. Here RNA polymerase is moving down the DNA template, unwinding the DNA ahead of it, and adding nucleotides one at a time (extending the 3′ OH)
As already mentioned, an RNA chain, complementary to the DNA template, is built by the RNA polymerase by the joining of the 5′ phosphate of an incoming ribonucleotide to the 3’OH on the last nucleotide of the growing RNA strand. Behind the RNA polymerase, the DNA template is rewound, displacing the newly made RNA from its template strand.
Termination: ‘Ending transcription’. Here the terminator sequences are recognized, the separation of the RNA molecule from the DNA template occurs and the enzyme ‘falls off’ as it is no longer needed.
Not unlike DNA Replication, the most elaborate step in transcription is Initiation which we will discuss first
6.6.1 Transcriptional Initiation
The events that comprise transcriptional initiation can be conceptually broken down into
1) Recognition of promoter 2) Unwinding of dsDNA and creation of bubble and 3) Initial synthesis and escape of RNA polymerase from the promoter.
Below is a summary of the events
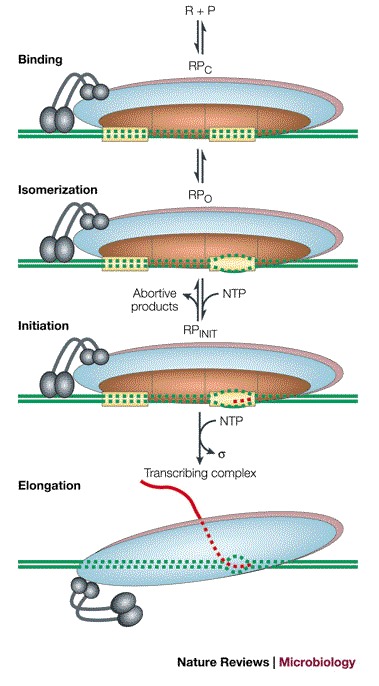
a. RNA polymerase holoenzyme binds to the promoter to form a closed complex; at this stage, there is no unwinding of DNA.
b. Closed complex is then converted to an “open” complex by the separation of the DNA strands to create a transcription bubble about 12-14 base-pairs long. The conversion of the closed complex to the open complex also requires the presence of the σ subunit. The section of promoter DNA that is within it is known as a ‘transcription bubble’. The transcription bubble is about 12-14 base-pairs long.
Role of sigma factor
- Initial specific binding to the promoter by sigma factors of the holoenzyme sets in motion conformational changes that result in the separation of the two strands of DNA and expose a portion of the template strand.
- The sigma protein consists of different subunits. Some portions of sigma interact with the −35 element. The duplex DNA just upstream of the −10 element (−17 to −13) interacts with other parts of the protein.
- The structure of the sigma unit is such that there is a recognition pocket for a nucleotide– the A−11(nt) base from the duplex DNA. This base gets flipped into its recognition pocket in σA2 is thought to be the key event in the initiation of promoter melting and the formation of the transcription bubble.
- Once the transcription bubble has formed and transcription initiates.
c. Abortive initiation: Once the open complex has formed, the DNA template can begin to be copied, and the core polymerase adds nucleotides complementary to one strand of the DNA. The polymerase adds several nucleotides while still bound to the promoter, and without moving along the DNA template. Initially, short pieces of RNA a few nucleotides long may be made and released, without the polymerase leaving the promoter.
Part of it is due to the contacts the sigma factor still has with the promoter.
d. Promoter Escape: After several abortive initiation attempts, the polymerase synthesizes an RNA molecule from 9 to 12 nucleotides in length, which allows the polymerase to transition to the elongation stage. The σ subunit also dissociates from the core enzyme which ‘breaks free’ or ‘escapes’ into the gene.
Nature Reviews Microbiology 2(1):57-65
DOI:10.1038/nrmicro787
Exercises
6.6.2 Transcriptional Elongation
The core polymerase can move along the template, unwinding the DNA ahead of it to maintain a transcription bubble of 12-15 base pairs and synthesizing RNA complementary to one of the strands of the DNA.
RNA polymerases are by themselves processive (with no need for additional apparatus) adding hundreds or thousands of bases to the growing RNA at about 20-50 nucleotides per second.
Unlike DNA polymerases there is no elaborate mechanism for proof-reading. Although research has shown that RNA polymerase is capable of a type of proofreading in the course of transcription.
When RNA polymerase incorporates a nucleotide that does not match the DNA template, it backtracks and cleaves the last two nucleotides.
Transcription Termination
As mentioned earlier, a sequence of nucleotides called the terminator is the signal to the RNA polymerase to stop transcription and dissociate from the template.
Some terminator sequences, known as intrinsic terminators, allow termination by RNA polymerase without the help of any additional factors, while others, called Rho-dependent terminators, require the assistance of a protein factor called rho (ρ).
How does the sequence of the terminator cause the RNA polymerase to stop adding nucleotides and release the transcript?
To understand this, it is useful to know that the terminator sequence precedes the last nucleotide of the transcript. In other words, the terminator is part of the end of the sequence that is transcribed.
Intrinsic terminators
These makeup about 50% of all terminators in prokaryotes and have two common features.
First, they contain inverted repeats, which are sequences of nucleotides on the same strand that are inverted and complementary. This sequence when transcribed into RNA can base-pair with each other to form a hairpin structure that contains a GC-rich run in the “stem” of the hairpin.
Second, adjacent to the inverted repeat is a stretch of 7- 9 Adenines. These when transcribed get converted to ‘U’s.
As RNA polymerase reaches and transcribes the terminator region the RNA will have a stem-loop structure. The secondary structure formed by the folding of the end of the RNA into the hairpin causes the RNA polymerase to pause!
Meanwhile, the run of U’s at the end of the hairpin permits the RNA-DNA hybrid in this region to come apart, because the base-pairing between A’s in the
DNA template and the U’s in the RNA is relatively weak.
This allows the transcript to be released from the DNA template and from the RNA polymerase.
Rho-dependent termination
Transcription termination factor Rho is an essential protein in E. coli first identified for its role in transcription termination at Rho-dependent terminators, and is estimated to terminate ~20% of E. coli transcripts. The rho gene is highly conserved and nearly ubiquitous in bacteria.
Rho is a helicase and consists of a hexamer of six identical monomers arranged in an open circle. The protein can separate the transcript from the template it is paired
As in intrinsic termination, rho-dependent termination requires the formation of a hairpin structure in the RNA that causes pausing of the RNA polymerase.
Meanwhile, rho binds to a region of the transcript called the rho utilization site (rut), a∼ cytidine-rich and poorly structured RNA sequence, and moves along the RNA till it reaches the paused RNA polymerase.
It then acts on the RNA-DNA hybrid, releasing the transcript from the template.
Remember to:
- Watch the Lecture videos that cover the material above. This will help to clarify or reinforce certain concepts if they were unclear.
- Complete the associated Lecture Quick checks
- Begin work on Problem Set.
References and Attributions
This chapter contains material taken from the following CC-licensed content. Changes include rewording, removing paragraphs and replacing with original material, and combining material from the sources.
1. Bergtrom, Gerald, “Cell and Molecular Biology 4e: What We Know and How We Found Out” (2020). Cell and Molecular Biology 4e: What We Know and How We Found Out – All Versions. 13.
https://dc.uwm.edu/biosci_facbooks_bergtrom/13
2. Works contributed to LibreTexts by Kevin Ahern and Indira Rajagopal. LibreTexts content is licensed by CC BY-NC-SA 3.0. The entire textbook is available for free from the authors at http://biochem.science.oregonstate.edu/content/biochemistry-free-and-easy
3. Flatt, P.M. (2019) Biochemistry – Defining Life at the Molecular Level. Published by Western Oregon University, Monmouth, OR (CC BY-NC-SA). Available at: https://wou.edu/chemistry/courses/online-chemistry-textbooks/ch450-and-ch451-biochemistry-defining-life-at-the-molecular-level/?preview_id=4919&preview_nonce=cca8f0ce36&preview=true

