Molecular Biology: From DNA to RNA to Protein
8 Eukaryotic Transcription and Regulation
Learning Objectives
- Describe the RNA polymerases of eukaryotes, locate them within the cell, and list the kinds of RNA they synthesize.
- Describe the three processes that commonly modify eukaryotic pre-mRNA.
- List the salient sequence elements of eukaryotic promoters. Contrast the nucleotide sequences and locations of the TATA box of eukaryotes and the -10 sequence of prokaryotes.
- What elements constitute a ‘core promoter’.
- Explain why eukaryotic promoters are more variable than bacterial promoters.
- Bacterial and eukaryotic gene transcripts can differ, in the transcripts themselves, in whether the transcripts are modified before translation, and in how the transcripts are modified. For each of these three areas of describe what the differences are.
- What are Enhancer sequences and how are they different from core promoter sequences?
- What does it mean that enhancers are position- and orientation-independent?
- What is combinatorial control?
- What role does the mediator play in transcription
- How do transcriptional repressors work? (In lecture videos)
- What purposes do capping and poly-A tail addition serve for eukaryotic mRNAs?
•Show the pathway for cap formation
• Answer at what stage of mRNA formation is the cap added to the RNA molecule. - Describe the basic assembly of PIC as deduced from in vitro experiments.
- Explain the key role of TATA-box-binding protein (TBP) in assembling active TFII transcription complexes.
- Describe the role of phosphorylation of the carboxy-terminal domain (CTD) of RNA polymerase II.
LEVEL UP (combines molecular bio methods introduced thus far, and others introduced later)
- Interpret reporter gene assay data for the identification of regulatory elements (all types including enhancers) in eukaryotic genes.
- Interpret data that leads to the identification of general transcription factors.
- Explain how mutations in regulatory regions of genes differ from mutations in the coding region.
8.1 Introduction
In Chapter 7 Introduction to Transcription, you learned about the basics of transcription (which is the same for prokaryotes and eukaryotes) as well as some differences between transcription in prokaryotes and eukaryotes. As a reminder, some differences are listed again below
- E. coli uses a single RNA polymerase enzyme to transcribe all kinds of RNAs while eukaryotic cells use different RNA polymerases to catalyze the syntheses of ribosomal RNA (rRNA), transfer RNA (tRNA), and messenger RNA (mRNA).
- In contrast to eukaryotes, some bacterial genes are part of operons whose mRNAs encode multiple polypeptides. Eukaryotic genes are not part of operons.
- Most RNA transcripts in prokaryotes emerge from transcription ready to use for translation! In eukaryotes, transcription and translation occur in different compartments (nucleus – cytoplasm)
- Eukaryotic transcripts synthesized as longer precursors undergo processing by trimming, splicing, or both!
Importantly eukaryotic DNA is wrapped up in chromatin proteins in a nucleus. Therefore the default state of transcription in eukaryotes is ‘off’!
The implication of this is that to begin transcription of any eukaryotic gene, there must be mechanisms to ‘activate it’. Eukaryotic transcription initiation cannot be separated from regulation!
This chapter consists of descriptions of key concepts and terms introduced in the lecture videos.
Research methodology and specific biomedical or relevant examples are highlighted in detail within the lecture videos.
8.2 Eukaryotic Cells Have Three Types of RNA Polymerase
RNA Polymerase I (Pol I) is responsible for the synthesis of the majority of rRNA transcripts, whereas RNA Polymerase III (Pol III) produces short, structured RNAs such as tRNAs and 5S rRNA. RNA Polymerase II (Pol II) produces all mRNAs and most regulatory and untranslated RNAs.
Did You Know?
The death cap mushroom produces a toxin α- Amanatin. The lethal effect of this toxin is due to its effect on RNA polymerases. The poison binds very tightly to RNA polymerase II and effectively prevents transcription.
The chemistry of RNA polymerization is identical in all types of organisms, and the three eukaryotic RNA polymerases are structurally related to E. coli RNA Polymerase; consist of homologs of 5 prokaryotic core subunits that form the same characteristic crab-claw shape in addition to other subunits.
In addition to homologs of the core subunits, there are many more polypeptides that make up the eukaryotic RNA polymerases.
One of the subunits of RNA Polymerase II possesses a unique CTD (carboxy-terminal domain) consisting of multiple repeats of a special heptameric (Hepta- 7) amino acid sequence Tyr-Ser-Pro-Thr-Ser-Pro-Ser that repeats itself.
In mammals, this domain consists of 52 repeats of the amino acid sequences. Serines in each repeat unit can be modified by the addition of a phosphate group, causing a substantial change in the properties of the polymerase.
The phosphorylation of the CTD of RNA polymerase plays an important role in transcription and mRNA processing.
8.3 Overview of Gene Expression (From DNA to Protein)
We focus on initiation by RNA pol II, the polymerase that produces all mRNAs and most regulatory and untranslated RNAs. Below (Figure 8.1) is a diagram of elements of the eukaryotic gene- that include all the sequences necessary to regulate transcription in addition to the protein-coding sections.
The structure of eukaryotic genes includes features not found in prokaryotes (Figure 8.1).
Eukaryotic genes typically have more regulatory elements to control gene expression compared to prokaryotes.
An additional layer of regulation occurs for protein-coding genes after the mRNA has been processed to prepare it for translation to protein.
Only the region between the start and stop codons encodes the final protein product. The flanking untranslated regions (UTRs) contain further regulatory sequences. The 3′ UTR contains a terminator sequence, which marks the endpoint for transcription and releases the RNA polymerase, and also contains sequences that regulate mRNA stability.
The 5’ UTR contained sequences that serve as landing pads for translational machinery (ribosome and other factors). In the case of genes for non–coding RNAs, the RNA is not translated but instead folds to be directly functional.
The most striking difference is the extent to which eukaryotic mRNA (pre-mRNA) is modified to produce mature mRNA ready for translation into protein.
These include:
– addition of a 5′ CAP at 5′ end of mRNA produced.
– splicing, the removal of the intron regions, and joining together of exons (the protein-coding portions)
– addition of a Poly A tail (polyadenylation) that is an inherent part of the termination mechanism.
Importantly most processing occurs while mRNA is being synthesized (co-transcriptional) and some soon after transcription (post-transcriptional). For example, the cap is added as soon as transcription has been initiated, splicing and editing begin while the transcript is still being made.
However to deal with all of these events together would be confusing, with too many different things being described at once.
We will therefore postpone mRNA processing until after we have talked about the Initiation of transcription. We will consider splicing completely separately after we discuss capping, elongation, and polyadenylation.
8.4 Details of Eukaryotic Transcription Initiation
As depicted in Figure 8.1 transcription starts downstream of the promoter and creates a transcript that begins with a 5’ untranslated region (5’UTR) followed by the coding region which may include multiple introns and ending in a 3’ untranslated region or 3‘UTR.
RNA Pol II gene transcription in eukaryotes is tightly regulated, controlled by a highly complex multicomponent machinery. A plethora of proteins, more than a hundred in humans, are organized in often very large multiprotein assemblies.
8.4.1 Eukaryotic Promoters
Eukaryotic promoters are more complex. They include all the sequences that are necessary for both initiation of transcription of a gene as well as regulatory sequences.
The promoter is located at the 5′ end of the gene and can be divided into the
CORE PROMOTER– which represents a minimal set of sequences necessary for assembly of the transcription machinery and transcription initiation. This allows for BASAL LEVELS of TRANSCRIPTION.
‘PROXIMAL PROMOTER ELEMENTS” – regulatory sequences next to the core promoter sequences.
While the assembly of the initiation complex can occur on core promoter sequences, almost all genes have additional proteins called Transcriptional Activators that bind to Proximal Promoter elements and ‘promote’ gene expression.
An alternative term used for these are “upstream promoter elements” or “upstream regulatory elements”
“ENHANCERS/SILENCER- DISTAL“- these sequences are located many thousands of base pairs away. The binding of different transcription factors, therefore, regulates the rate of transcription initiation at different times and in different cells.
Enhancer and Silencer sequences dictate when (developmental stage) and where (what tissue) a gene gets expressed.
Core Promoter Sequences:
The core promoter is a region encompassing approx 50 base pairs (bp) upstream and aprox 50 bp downstream of the TSS. It includes or encompasses the TSS!
First, it is important to note that not all eukaryotic genes look alike! There is no exact set sequence or a minimum number of core promoter sequences. Most genes have some combinations of elements, and scientists are still identifying core promoter consensus sequences.
Below are some “typical core promoter consensus ” sequences.
A TATA box (consensus 5′-TATAAA-3′,) –about 25-35 base pairs upstream of the start of transcription (+1). (Note this is not the same as the sequence found in prokaryotes but is similarly A-T-rich to facilitate the opening of DNA and formation of transcription bubble)
Initiator (Inr) sequence located around nucleotide +1 (the TSS)
DPE (downstream promoter element): is a common component of RNA polymerase II promoters that do not contain a TATA box (TATA-less promoters).
8.4.2 Role of General Transcription Factors
A key difference between the initiation of transcription in E. coli and eukaryotes is that eukaryotic polymerases do not directly recognize their core promoter sequences.
The core promoter sequences described above are recognized by a set of proteins called general (or basal) transcription factors. These proteins are not part of the RNA polymerase II complex.
These general transcription factors are found in all eukaryotes, suggesting that the fundamentals of transcription are conserved in higher organisms.
These proteins are identified as TFNX, where N is a roman numeral I, II, or III (signifying the polymerase) and X is a letter.Therefore TF-II – means Basal Transcription Factor for RNA Pol II.
Note:
The bulk of the work identifying general transcription factors as well as the order of assembly was done using genes with promoters that have the TATA box and deduced by in vitro experiments.
Establishing the Pre-Initiation Complex
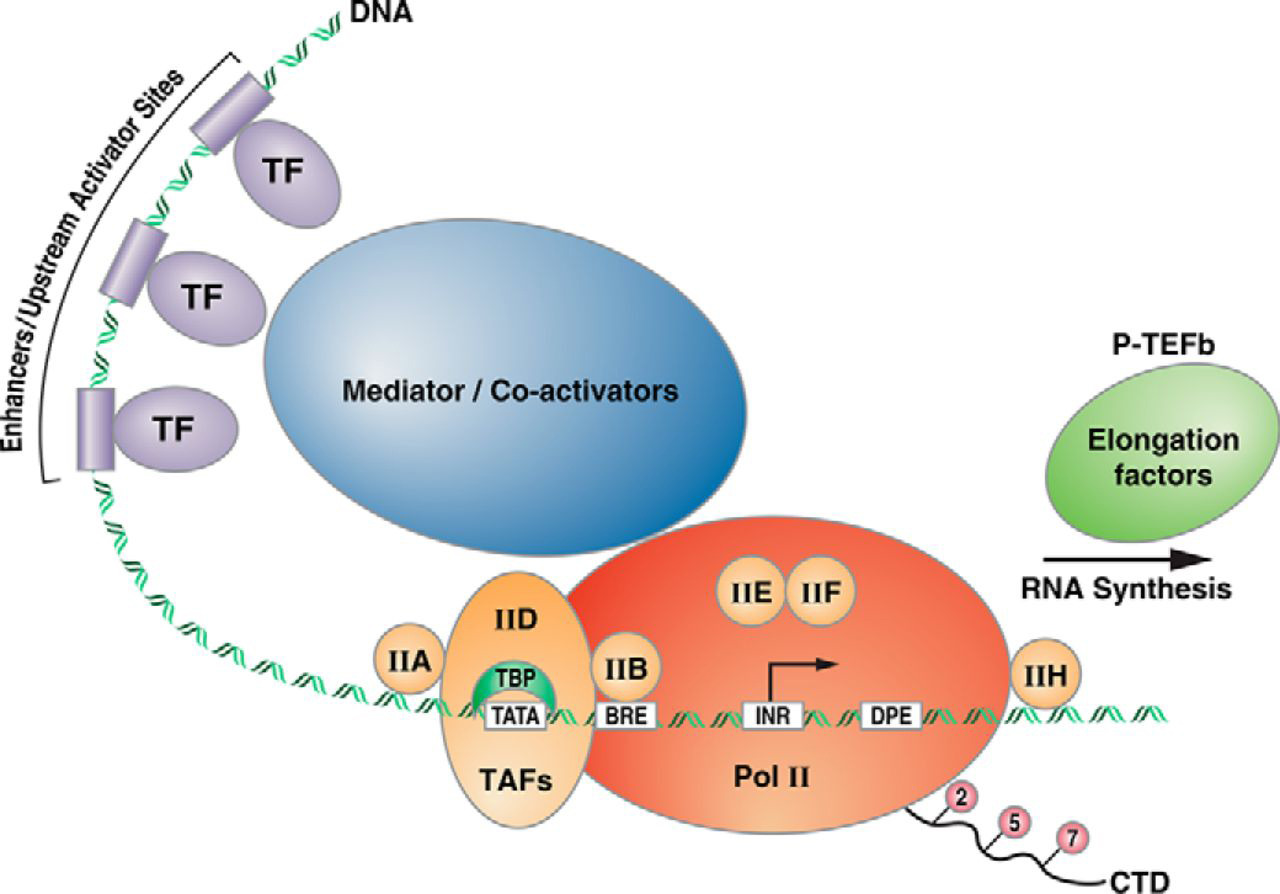
In order for transcription to occur RNA polymerase II needs to be recruited to the appropriate location- around the transcription start site, on the core promoter. The first steps in eukaryotic transcription involve the regulated assembly of the general transcription factors (GTFS).
These proteins serve as a platform for RNA polymerase II recruitment.
The GTFs include the factors TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH, RNA polymerase (RNA pol II).
We will only focus on the functions of 2 GTFs: TFII-D an TFII-H
1. TFII-D is a large multi-subunit protein that includes the TATA-binding protein.
As the name “TATA-binding protein” suggests: TBP binds to the TATA box. X-ray crystallography studies of TBP show that it has a saddle-like shape that wraps partially around the double helix. (Chasman DI, et al)
2. Binding of TFIID to the core promoter (via TBP) is followed by the recruitment of additional GTFs in a step-wise fashion and eventually RNA pol II.
The combination of all the GTFs along with RNA Pol II is the Pre-initiation Complex (PIC)
PIC first adopts an inactive state, the “closed” complex, which is incompetent to initiate transcription. This complex is ‘poised for transcription’.

Abortive Initiation, Promoter Clearance, and Elongation
TFII-H also a multisubunit protein plays a key role in the transition from ‘closed to open complex’.
This protein has several components that come together to have 2 enzymatic activities:
1) ATP-dependent helicase type activity- that opens up about 11 to 15 base pairs around the transcription start site leading to forming the transcriptional bubble.
Abortive transcription- once the RNA polymerase binds, it can begin to assemble a short stretch of RNA. This must be followed by promoter clearance, in order to move down the template and elongate the transcript.
2) TF II H- Kinase activity: adds phosphates onto the C-terminal domain (CTD) of the RNA polymerase II. ( specifically certain amino acids within the CTD (C-terminal domain) of RNA polymerase II get phosphorylated)
[ Terminology alert: Kinases are the name given to a class of enzymes that catalyze the transfer of a phosphate group from ATP to proteins. Commonly modified amino acids include Serines, Threonines, Tyrosines]
This phosphorylation appears to be the signal that releases the RNA polymerase from the basal transcription complex and allows it to move forward on the template, building the new RNA as it goes
After the departure of the polymerase, at least some of the GTFs detach from the core promoter.
8.4.3 Other Regulatory Sequences
Fundamentally, a key difference with bacterial transcription is that the pre-initiation complexes do not assemble efficiently and the basal rate of transcription initiation is therefore very low, regardless of how ‘strong’ the promoter is.
As was discussed in the outlining the structure of eukaryotic promoter, in most eukaryotic genes to achieve effective initiation, the formation of the complex must be activated by additional proteins.
Any protein that stimulates transcription initiation is called a Transcriptional Activator. These proteins bind either the Promoter Proximal Elements or Enhancer/Silencer sequences. This binding is sequence-specific.
Promoter Proximal Elements
Are several different consensus sequences to which different regulatory transcription factors can bind.
In different promoters, transcription factor binding sites are mixed and matched in different combinations. Each promoter is regulated by a unique combination of transcription factors.
The binding of transcription factors to the consensus sequences in the regulatory promoter affects the assembly or stability of the basal transcription apparatus at the core promoter.
Example: Red blood cell development
An example of the former is the upstream element AACCAAT and its associated transcription factor, CP1. Another transcription factor, Sp1, is similarly common and binds to a consensus sequence of ACGCCC.
Both are used in the control of the beta-globin gene, along with more specific transcription factors, such as GATA-1, which binds a consensus AAGTATCACT and is primarily produced in blood cells.
CP1 is found in many types of cells. GATA-1 is present in only a few types of cells including red blood cells; therefore is thought to contribute to the cell-type specificity of β-globin gene expression
This illustrates another option found in eukaryotic control that is not present in prokaryotes: tissue-specific gene expression.
Response Elements
In addition, many genes have common regulatory elements called ‘RESPONSE ELEMENTs’. These response elements are binding sites for Transcriptional Activators and enable transcription initiation to respond to general signals from outside of the cell:
Examples:
- the cyclic AMP response module CRE (consensus 5 -WCGTCA-3 ), recognized by the CREB activator
- heat-shock module (HSE) (consensus 5 -CTNGAATNTTCTAGA-3 ), recognized by HSP70 and other
activator - steroid- hormone response element [Glucocorticoid Response Element, Estrogen Receptor Element]
Enhancers and Silencers
Enhancers are regulatory elements that stimulate the transcription of distant genes. Silencers inhibit transcription.
Both regulate transcription over long distances in a position- and orientation-independent manner.
Enhancers are transcription activator binding sites grouped in units. Multiple enhancers enable a gene to respond differently to different combinations of activators.
This arrangement gives cells, in a developing organism, exquisitely fine control over their genes in different tissues or at different times!
The ‘looping of DNA’ between the enhancer sites and the core promoter region helps proteins bound to the enhancer interact with the transcriptional apparatus.
Concepts in Context
Watch the short film The Making of the Fittest: Evolving Switches, Evolving Bodies. Pay close attention to how the switches regulate the expression of the Pitx1 gene in stickleback embryos as well as the ‘Reporter Gene Assay’ used.
REMEMBER: Don’t forget to complete the associated assignment in CANVAS.
8.4.4 Mediator Complex
Experimental studies of transcription in vitro showed that in addition to the GTFs, another multisubunit complex mediates communication between activating TFs (at enhancer and upstream activator sequences) and the GTFs and RNA pol II, hence the name “Mediator” for this complex. (Ref)
According to current gene activation models, the Mediator complex forms a physical bridge between proteins bound to distant regulatory regions and promoters, and transcription machinery at the core promoter.
The mediator is a huge complex of 25 to 30 subunits with a mass of more than 1-MDa.
A picture of transcription initiation that includes all the elements is shown in Figure 8.4 below
8.5 How Transcription factors Work
Experiments using recombinant proteins showed that transcriptional activators are modular containing 2 domains.
[Recall the function of a protein domain from Chapter 1]
DNA binding domain: which contacts the regulatory sequences
The DNA binding domains fall into one of four representative families that are distinct structurally.
Activation domain: responsible for ‘activation’ or recruitment of transcriptional machinery.
Transcription factors often work as dimers.
In general, most regulatory transcription factors do not bind directly to the RNA polymerase
Ways in which Transcriptional Activators influence transcription include
- Influence the PIC at the promoter directly via TF-II D or indirectly via the mediator
- Influence the chromatin structure!
The main way in which this can be achieved as was discussed in Chapter 4 is
- Covalent modification of histones
- ATP-dependent chromatin remodeling.
Both of these activities are present in many transcription factors!
8.5 Bringing it all together
Overall the picture of transcription initiation then is less of an ON or OFF but that of fine-tuning.
Transcription initiation in vivo requires the presence of transcriptional activator proteins (coded by gene-specific transcription factors). These proteins bind to specific short sequences in DNA (enhancers).
A typical eucaryotic gene has many activator proteins, which together determine its rate and pattern of transcription. Sometimes acting from a distance of several thousand nucleotide pairs, these gene regulatory proteins help RNA polymerase, the general factors, and the mediator all assemble at the promoter.
In addition, activators attract ATP-dependent chromatin-remodeling complexes and histone acetylases.
Each individual gene transcription can be adjusted in amount based on the tissue type, developmental stage, and biochemical condition.
Factors like the number of sequences, types of enhancers, presence or absence of transcriptional factors, co-activators, or repressors all dictate the final outcome of transcription initiation.
8.6 Relevant Biological Concepts
The mixing and match of these regulatory sequences is the principle behind 2 important features of eukaryotic transcriptional regulation
Coordinated Gene Regulation and Combinatorial Control
The presence of the same response element in different genes allows a single stimulus to activate multiple genes by virtue of binding to a single transcriptional regulator.
This phenomenon is also behind the success of gene expression patterns during development, which results in establishing cell fate.
Similarly, the Transcription factors and other proteins that bind to regulatory sites on DNA provide RNA polymerase, access to specific genes. Therefore a given regulatory protein can have different effects, depending on what other proteins are present in the same cell. This phenomenon is combinatorial control.
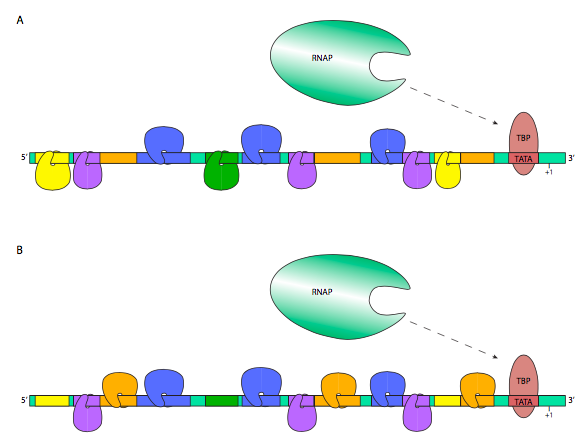
An illustrative example is shown below. In the figure, the transcription factors hanging downward are representative of inhibitory TFs, while those riding upright on the DNA are considered activating. Thus, the RNA polymerase in (A) has a lower probability of transcribing this gene, while the RNAP in (B) is more likely to, perhaps because the TF nearest the promoter interacts with the RNAP to stabilize its interactions with TFIID.
In this way, the same gene may be expressed in very different amounts and at different times depending on the transcription factors that are present in a particular cell type.
Molecular Biology in the News: Tissue Engineering
Molecular mechanisms that create and maintain specialized cell types depend on -‐Combinatorial gene control. Combinations of master transcription regulators specify cell types by controlling the expression of many genes.
This discovery is the basis behind induced pluripotent stem (iPS) cells- the ability to take specialized cells (like skin or fibroblasts) and reprogram them to become immature cells. The addition of four genes, encoding transcription factors can induce these cells to become pluripotent stem cells, i.e. immature cells that are able to develop into all types of cells in the body.
The Nobel Prize in Physiology or Medicine 2012 was awarded for this discovery and has implications broader biomedical implications- including creating organs within a lab for organ donation.
REMEMBER: Complete the reading associated with this within your module.
Learning Objectives: You should be able to:
- Describe the three processes that commonly modify eukaryotic pre-mRNA.
- Bacterial and eukaryotic gene transcripts can differ, in the transcripts themselves, in whether the transcripts are modified before translation, and in how the transcripts are modified. For each of these three areas of contrast, describe what the differences are.
- Draw the structure of the end of a typical eukaryotic mRNA. Outline the reactions required to cap the primary transcript.
- Describe the events leading to the production of mRNA with a poly(A) tail.
- What purposes do capping and poly-A tail addition serve for eukaryotic mRNAs?
- Answer at what stage of mRNA formation is the cap added to the RNA molecule.
- Explain how Polyadenylation and transcription termination are linked.
- List the components of the posttranscriptional machinery recruited through interactions with the CTD of RNA polymerase II.
8.7 Transcription Elongation and Termination- mRNA Processing
After initiation, the mechanics of transcription elongation are similar to that in Prokaryotes, however, a big difference is the modification of the mRNA as it emerges from the RNA pol II enzyme.
The first modification occurs at the 5′ end
5’ end-capping.
Once the 5’ end of a nascent RNA extends free of the RNAP II approximately 20-30 nt, it is ready to be capped by a 7-methylguanosine structure.
It consists of a guanine nucleotide connected to mRNA via an unusual 5′ to 5′ triphosphate linkage (Figure below). This guanosine is methylated on the 7 position directly after capping in vivo by a methyltransferase.
It is referred to as a 7-methylguanylate cap, abbreviated m7G.
The process involves three steps.
First, RNA triphosphatase removes the 5’-terminal triphosphate group.
Second, Guanylation by GTP is catalyzed by a capping enzyme, forming an unusual 5’-5’ “backward” bond between the new guanine and the first nucleotide of the RNA transcript.
Finally, guanine-7-methyltransferase methylates the newly attached guanine.
This 5’ “cap” has many functions.
- serves as a recognition site for transport of the completed mRNA out of the nucleus and into the cytoplasm
- Prevention of degradation by exonucleases
- Promotion of translation (see ribosome and translation)
3’ end Polyadenylation and termination
The 3′ end of the gene (within the 3’UTR) is the signature sequence for signaling the end of transcription and polyadenylation.
It consists of a Poly A site flanked by a polyadenylation signal (AATAAA) and a downstream element that is GT-rich.
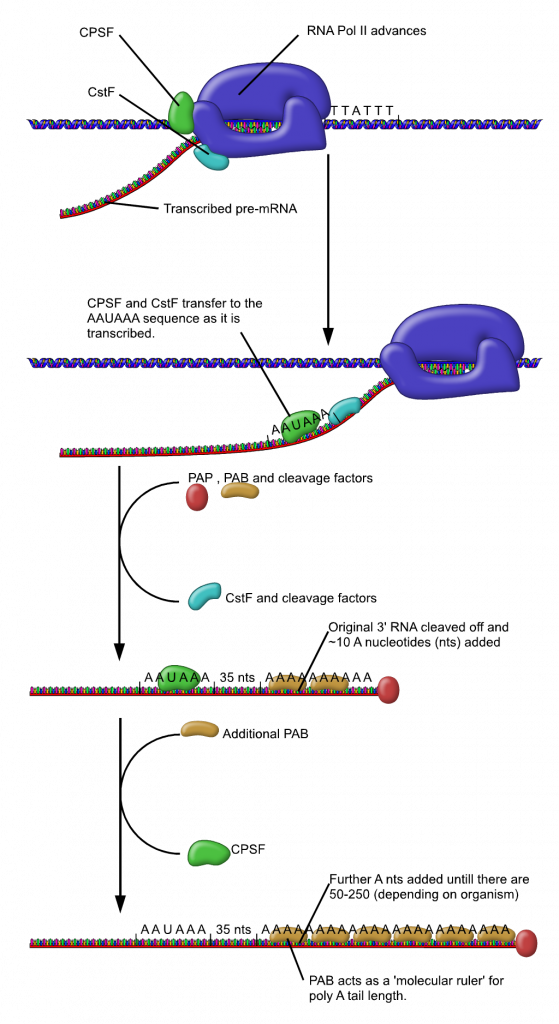
As the transcriptional machinery marches along the gene it will eventually transcribe this region generating within the transcribed mRNA the consensus AAUAAA sequence and the downstream element which will be a GU-rich sequence.
A protein complex called CPSF (the cleavage and polyadenylation specificity factor, CPSF) recognizes the poly-A signal. It has endo-nuclease activity and cuts the pre-mRNA between the AAUAAA consensus sequence and the GU-rich sequence, leaving the AAUAAA sequence on the pre-mRNA and a free 3′ OH.
Note: This releases the mRNA from the transcribing machinery!
An enzyme called poly-A polymerase then adds a string of approximately 200 Adenine residues, called the poly-A tail.
NOTE: The poly A tail is NOT a part of coded information of the gene but added post-transcriptionally !
Evidence suggests that the Poly A tail influences the efficiency of translation. The poly-A tail also has an effect on the stability of transcripts in the cytoplasm.
Splicing
The third and most complicated modification to newly-transcribed eukaryotic RNA is splicing. Splicing is the process by which the non-coding regions, known as introns, are removed, and the coding regions, known as exons, are connected together. We will be discussing the mechanism of splicing separately, although it is useful to introduce it here because splicing is also occurring during transcription!
Role of CTD of RNA Polymerase in mRNA processing
That transcription and mRNA processing are coupled is highlighted by the fact that proteins utilized for capping, splicing, and polyadenylations are recruited to the CTD of RNA pol II!
Recall that the CTD consists of multiple repeats of a special heptameric (Hepta- 7) amino acid sequence Tyr-Ser-Pro-Thr-Ser-Pro-Ser. In particular, the Serines may be phosphorylated in the various repeats. This occurs in a sequential manner and creates a signature (like a code) for many of the processing proteins to bind to the tail!
The RNA Pol-II enzyme physically carries the processing enzymes with it- and they get deployed as needed!
Check your understanding
Animation of Eukaryotic Transcription
References and Attributions
This chapter contains material taken from the following CC-licensed content. Changes include rewording, removing paragraphs and replacing with original material, and combining material from the sources.
1. Bergtrom, Gerald, “Cell and Molecular Biology 4e: What We Know and How We Found Out” (2020). Cell and Molecular Biology 4e: What We Know and How We Found Out – All Versions. 13.
https://dc.uwm.edu/biosci_facbooks_bergtrom/13
2. Works contributed to LibreTexts by Kevin Ahern and Indira Rajagopal. LibreTexts content is licensed by CC BY-NC-SA 3.0. The entire textbook is available for free from the authors at http://biochem.science.oregonstate.edu/content/biochemistry-free-and-easy
3. Flatt, P.M. (2019) Biochemistry – Defining Life at the Molecular Level. Published by Western Oregon University, Monmouth, OR (CC BY-NC-SA). Available at: https://wou.edu/chemistry/courses/online-chemistry-textbooks/ch450-and-ch451-biochemistry-defining-life-at-the-molecular-level/?preview_id=4919&preview_nonce=cca8f0ce36&preview=true
4. “Eukaryotic Transcriptional Regulation” by E. V. Wong, LibreTexts is licensed under CC BY-NC-SA
Other References
Chasman DI, Flaherty KM, Sharp PA, Kornberg RD. Crystal structure of yeast TATA-binding protein and model for interaction with DNA. Proc. Natl Acad. Sci. USA. (1993);90:8174–8178. [PMC free article]

